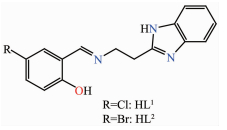
 Figure Scheme 1.
Structures of Schiff base ligands HL1 and HL2
Figure Scheme 1.
Structures of Schiff base ligands HL1 and HL2

两个基于苯并咪唑席夫碱的镍(Ⅱ) 配合物的合成、晶体结构和抑菌活性
English
Two Ni(Ⅱ) Complexes of Schiff Base Ligands Containing Benzimidazole Ring: Syntheses, Crystal Structures and Antibacterial Properties
-
Key words:
- Schiff base
- / Ni(Ⅱ) complex
- / crystal structure
- / antibacterial activity
-
0 Introduction
For decades, Schiff bases and their transition metal complexes have attracted a lot of attention due to their wide biological applications[1-5]. It is obvious that in Schiff bases the azomethine linkage (C=N) is an important structural requirement for biological activity[6]. Metal complexes containing salicylidene Schiff bases have been of great interest due to their diverse roles in magnetic, luminescence[7-8] and phar-macological properties[9]. It was also reported that sali-cylidene Schiff base with halogen atoms in the sali-cylaldehyde moiety had greater biological activities, like antibacterial and antifungal activities[1, 10-11]. Ben-zimidazole and their derivatives have received consi-derable attention in recent years due to their wide variety of biological activities including antimicroa-bial[12], antifungal[13], antitumor[14], antiviral[15] etc. However, literature survey shows that study on metal complexes of benzimidazole-derived salicylidene Schiff bases is not rich. Until now, a limited number of Cu(Ⅱ), Ni(Ⅱ) and Ⅴ(Ⅳ, Ⅴ) complexes with the above type of Schiff base derivatives have been reported[16-24]. On the other hand, much attention has been paid to nickel complexes of Schiff base derivatives because such complexes have various applications, such as, as antibacterial agents, as fungicide agents, in the treat-ment of cancer and for other biological activities[25-27].
Herein, we report the syntheses, spectral characterizations and crystal structures of two Ni(Ⅱ) complexes by using two benzimidazole-derived Schiff base ligands HL1 and HL2 (Scheme 1). Moreover, antibacterial activities of the two ligands and two complexes were also investigated against S. aureus (Gram-positive) and E. coli (Gram-negative) bacteria.
1 Experimental
1.1 Material and physical measurement
2-Aminoethyl-1H-benzimidazole dihydrochloride was prepared by the method reported by Ceson et al.[28]. All of the other reagents were used as received. Microanalyses (C, H, N) were carried out using a Perkin-Elmer 240C analyzer. The infrared spectra in KBr pellets were obtained on a ThermoFisher Nicolet 6700 spectrometer in the 4 000~400 cm-1 region. Electronic spectra were carried out in methanol solvent on a Shimadzu UV-2550 spectrophotometer. 1H NMR spectra were taken in CDCl3 on a Bruker Avance 500MHz spectrometer at room temperature with tetramethylsilane as the internal standard.
1.2 Syntheses of the ligands
1.3 Syntheses of the complexes
1.4 X-ray crystallography
For the structure determination, suitable single crystals with dimensions of 0.30 mm×0.25 mm×0.17 mm (1) and 0.21 mm×0.19 mm×0.15 mm (2), respe-ctively, were mounted on a Bruker Smart 1000 CCD diffractometer, fine focus sealed tube equipped with graphite-monochromatized Mo Kα radiation (λ=0.071 073 nm) at 293(2) K. Semi-empirical absorption corrections were applied using SADABS program[29]. All the structures were solved by direct method, followed by full-matrix least-squares refinements on F2 with anisotropic displacement parameters for all non-hydrogen atoms using the programs SHELXS-97 and SHELXL-97[30]. All the hydrogen atoms were placed in their calculated positions and refined riding with their carrier atoms. The relevant crystal data and structure refinement for 1 and 2 are collected in Table 1. Selected bond lengths and bond angles are presented in Table 2.
 Table 1. Crystallographic data and refinement summary for the 1 and 2
Table 1. Crystallographic data and refinement summary for the 1 and 21 Ni1-N1 0.208 6(3) Ni1-N1ⅰ 0.208 6(3) Ni1-N2 0.208 2(3) Ni1-N2ⅰ 0.208 2(3) Ni1-O1 0.203 6(2) Ni1-O1ⅰ 0.203 6(2) O1ⅰ-Ni1-O1 94.36(13) N2ⅰ-Ni1-N1ⅰ 88.61(10) O1ⅰ-Ni1-N2ⅰ 174.77(10) N2-Ni1-N1ⅰ 97.13(10) O1-Ni1-N2ⅰ 87.75(10) O1ⅰ-Ni1-N1 87.77(10) O1ⅰ-Ni1-N2 87.75(10) O1-Ni1-N1 86.69(10) O1-Ni1-N2 174.77(10) N2ⅰ-Ni1-N1 97.13(10) N2ⅰ-Ni1-N2 90.55(15) N2-Ni1-N1 88.61(10) O1ⅰ-Ni1-N1ⅰ 86.69(10) N1ⅰ-Ni1-N1 171.85(15) O1-Ni1-N1ⅰ 87.77(10) 2 Ni1-N1 0.208 0(4) Ni1-N1ⅰ 0.208 0(4) Ni1-N2 0.208 9(4) Ni1-N2ⅰ 0.208 9(4) Ni1-O1 0.203 7(3) Ni1-O1ⅰ 0.203 7(3) O1ⅰ-Ni1-O1 94.79(18) N1ⅰ-Ni1-N2ⅰ 88.88(14) O1ⅰ-Ni1-N1ⅰ 86.57(13) N1-Ni1-N2ⅰ 97.56(10) O1-Ni1-N1ⅰ 87.23(13) O1ⅰ-Ni1-N2 87.43(13) O1ⅰ-Ni1-N1 87.23(13) O1-Ni1-N2 174.84(14) O1-Ni1-N1 86.57(13) N1ⅰ-Ni1-N2 97.56(14) N1ⅰ-Ni1-N1 170.8(2) N1-Ni1-N2 88.88(14) O1ⅰ-Ni1-N2ⅰ 174.84(14) N2ⅰ-Ni1-N2 90.7(2) O1-Ni1-N2ⅰ 87.43(13) Symmetry transformations used to generate equivalent atoms: ⅰ-x+1, y, -z+3/2 for 1; ⅰ-x+1, y, -z+1/2 for 2 Table 2. Selected bond lengths (nm) and bond angles (°) for 1 and 2CCDC: 1401182, 1; 1401183, 2.
1.5 Antibacterial activity test
The in vitro antimicrobial activity of Schiff base HL1 and HL2 and their respectively complexes 1~2 were studied against Staphylococcus aureus (as Gram positive bacteria) and Escherichia coli (as Gram negative bacteria) by the standard disc diffusion method[31]. The same procedure[32] was followed for the determination of zone inhibition of all the compounds against standard controls.
1.2.2 Synthesis of N-(Benzimidazol-2-ylethyl)-5-bromosalicylideneimine (HL2)
The preparation of HL2 followed the same proce-dure described for HL1 except that 5-bromosalicylald-ehyde was used instead of 5-chrolosalicylaldehyde.The ligand precipitated as a yellow solid. Yield: 90%. m.p. 191~193 ℃. IR (KBr pellet, cm-1): 2 840, 1 637, 1 415, 1 273, 752. 1H NMR (500 MHz, CDCl3, ): δ 3.29~3.34 (t, 2H, -CH2-), 4.12~4.17(t, 2H, -CH2-), 6.82~7.71(m, 7H, Ar-H), 8.29(s, 1H, -CH=N-), 13.22(b, 1H, OH). UV-Vis (methanol, λmax / nm): 274, 281, 330. Anal. Calcd. for C16H14BrN3O (%): C, 55.83; H, 4.10; N, 12.21; S, 18.7. Found (%): C, 55.60; H, 4.21; N, 12.34.
1.2.1 Synthesis of N-(benzimidazol-2-ylethyl)-5-chlorosalicylideneimine (HL1)
HL1 was synthesized by a condensation reaction between 2-aminoethyl-1H-benzimidazole dihydrochlo-ride (1.335 g, 5 mmol), previously neutralized with K2CO3 (0.83 g, 6 mmol), and 5-chlorosalicylaldehyde (0.605 g, 5 mmol) in 25 mL of methanol. The mixture was stirred at room temperature for two hours and then a yellow precipitate was obtained which was filtered off and washed with cold MeOH and then dried in air. Yield: 96%. m.p. 83~85 ℃. IR (KBr pellet, cm-1): 2 839, 1 637, 1 416, 1 274, 750. 1H NMR (500 MHz, CDCl3): δ 3.29~3.32(t, 2H, -CH2-), 4.12~4.15 (t, 2H, -CH2-), 6.86~7.55(m, 7H, Ar-H), 8.28(s, 1H, -CH=N-), 13.12(b, 1H, OH). UV-Vis (methanol, λmax / nm): 274, 281, 328. Anal. Calcd. for C16H14ClN3O (%): C, 64.16; H, 4.71; N, 14.03. Found (%): C, 64.30; H, 4.57; N, 14.12.
1.3.1 Synthesis of [Ni (L1)2]·2H2O (1)
Schiff base HL1 (0.060 g, 0.2 mmol) was dissolved in MeOH (10 mL) and 0.028 mL NEt3 was added. A solution Ni (ClO4)2·6H2O, (0.036, 0.1 mmol) in MeOH (10 mL) were then added with stirring at room temperature. The resulting green solution was stirred for two hours. Green yellow crystals suitable for X-ray structural analysis were obtained by slow evaporation of the solvent after several days. Yield: (0.055 g, 80%). IR (KBr pellet, cm-1): 3 656, 2 901, 1 622, 1 526, 1 458, 1 385, 1 325, 750. Anal. Calcd. for C32H30Cl2N6NiO4(%): C, 55.52; H, 4.37; N, 12.14. Found (%): C, 55.64; H, 4.43; N, 12.35.
1.3.2 Synthesis of [Ni (L2)2]·2H2O (2)
The preparation of complex 2 follows the same procedure as that of 1, except that HL2 (0.079 g, 0.2 mmol) was used. Yield: (0.065 g, 84%). IR (KBr pellet, cm-1): 3 649, 2 903, 1 620, 1 523, 1 458, 1 382, 1 327, 748. Anal. Calcd. for C32H30Br2N6NiO4(%): C, 49.20; H, 3.87; N, 10.76. Found (%): C, 49.31; H, 3.63; N, 10.51.
2 Results and discussion
2.1 Syntheses and characterization
The ligands HL1 and HL2 were prepared by the direct condensation reaction of 2-aminoethyl-1H-benzimidazole dihydrochloride, previously neutralized with K2CO3, with 5-chlorosalicylaldehyde and 5-bromosalicylaldehyde (molar ratio 1:1) in methanol, respectively. The metal complexes 1~2 were obtained by the reaction of corresponding ligands with Ni (ClO4)2 ·6H2O in a ratio of 2:1 (nligand / nNi) in methanol medium. The two complexes are air stable and soluble in EtOH, MeOH, DMSO and DMF.
Several medium intensity bands spreading between 3 300 and 2 300 cm-1 in the two complexes indicate that the NH groups of the benzimidazole rings are involved in hydrogen bonding with other electronegative atoms[21]. Bands at 3 656 and 3 649 cm-1 are assigned as ν(O-H) stretching vibrations of the hydrate water molecules in 1 and 2, respectively. The IR spectra of the two Schiff bases showed a very sharp and strong C=N stretching vibration around 1 637 cm-1. For the two complexes the same band was observed around 1 622 cm-1, which was shifted towards lower wavelength, suggesting coordination through the azomethine nitrogen atom of the ligands. The electronic spectra of the free Schiff base ligands and the two complexes in methanol were measured at room temperature. UV bands around 275 and 283 nm are observed for the free Schiff bases due to the π-π* transition of the benzimidazole group. In complexes 1 and 2 these bands blue-shift, indicating clear evidence of the ring nitrogen coordination to metal centers[33]. For the free Schiff bases the bands at 326 nm for HL1 and 330 nm for HL2, respectively, are characteristic of the n-π* transition of the azomethine linkage. On complexation, this band shifts to a longer wavelength in 1 and 2 (Δ=50 nm for both two complexes)[34]. In the visible region, the two complexes show a broad absorption band appears at λmax value of 640 nm, suggesting a distorted octahedral arrangement around the metal ions.
2.2 Crystal structures of the complexes
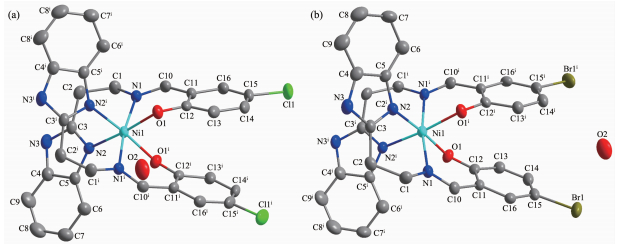
X-ray diffraction studies reveal that both complexes 1 and 2 crystallize in monoclinic system with C2/c space group. As shown in Fig. 1, in complexes 1 and 2, each complex molecule consists of a monomeric [Ni (L)2] unit with two solvent water molecules. The hexa-coordinated Ni(Ⅱ) center is bonded to two tridentate Schiff base ligands. Each ligand molecule offers deprotonated phenolic oxygen, imine nitrogen and benzimidazole nitrogen atom as the coordination sites providing a NiN4O2 chromophore. The geometry around Ni(Ⅱ) in 1 and 2 can be best described as a distorted octahedron, where the equatorial plane is constructed by two benzimidazole nitrogen atoms and two phenolic oxygen atoms, and the two axial sites being occupied by two imine nitrogen atoms. The average Ni-N and Ni-O distances are 0.208 4 and 0.203 7 nm, respectively, which are comparable to those reported for the similar Schiff base complexes of Ni(Ⅱ)[35]. The three trans-angles at nickel(Ⅱ) vary from 170.8(2)° to 174.84(14)° while the cis-angles are in the range of 86.57(13)~97.56(10)°, being deviating significantly from 180° and 90°, res-pectively, indicating the coordination geometry in com-plexes 1 and 2 is distorted from a regular octahedron.
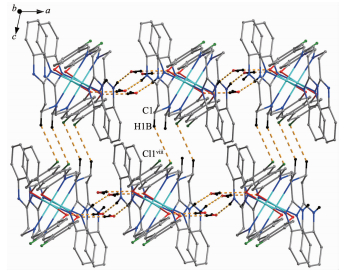
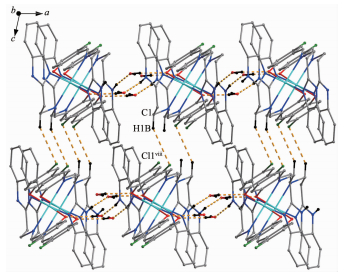
In complexes 1 and 2, the water molecules are involved in hydrogen bonds acting as both acceptors and donors leading to the formation of intermolecular interaction network. Acting as an acceptor, the O2 atom of the water molecule is engaged in the N3-H3…O2# hydrogen bonds to the H atom on the benzimidazole N atom. On the other hand, as a donor, the O2 atom is engaged with O1 atom of the phenolic group and the N2 atom of the benzimidazole ring through O2-H…O1# and O2-H…N2# hydrogen bonds, respectively, where # refers to the different L ligand. These hydrogen bonds link the complex 1 and 2 into a 2D structure (Fig. 2a and 2b). Besides, complex 1 was further linked into a 3D structure by the inter-molecular C1-H1B…Cl1 hydrogen bonds between a CH2 group bound to the imino moiety of L1 ligand and an adjacent Cl1 atom (Fig. 3). The bond parameters of hydrogen bonds are listed in Table 3.
D-H…A d(D-H)/nm d(H…A)/nm d(D…A)/nm ∠DHA/(°) 1 N3-H3 …02ⅱ 0.086 0.185 7 0.270 4 168.11 02-H2E …N2 0.085 0.248 5 0.322 6 146.28 02-H2F …01ⅲ 0.085 0.191 2 0.264 8 144.09 C1-H1B …C11ⅷ 0.097 0.292 4 0.379 6 150.00 2 N3-H3 …02ⅱ 0.086 0.185 0 0.269 9 169.07 02-H2E …01ⅲ 0.085 0.191 9 0.265 3 143.87 02-H2F …N2ⅳ 0.085 0.249 0 0.323 2 146.31 Symmetry codes: ⅱ-x+3/2, y-1/2, -z+3/2; ⅲ-x+1, y, -z+3/2; ⅷ-x+1, -y+1, -z+1 for 1; ⅱ x, y-1, z; ⅲ x+1/2, y+1/2, z; ⅳ-x+3/2, y+1/2, -z+1/2 for 2 Table 3. Hydrogen bond parameters in 1 and 22.3 Antibacterial activity of the compounds
The antibacterial activity of Schiff base ligands, HL1 and HL2 and their two Ni(Ⅱ) complexes were studied against one Gram-positive (S. aureus) and one Gram-negative (E. coli) bacterial strains and the results were showed in Table 4. The two Schiff base ligands are active against S. aureus and E. coli at different concentrations. HL1 exhibits greater antibacterial activity against E. coli whereas HL2 shows higher zone against S. aureus. The antibacterial activity of Schiff base ligands may be due to the presence of azomethine group as well as the presence of the hydroxyl and benzimidazole rings, all of which may play a significant role in the antibacterial activity[6, 10-12].
Compound Dose/(μg·mL-1) Inh1b1t1on zone diameter/mm S. aureus E. coli HL1 125 9 13 250 11 14 500 12 16 HL2 125 10 11 250 12 12 500 13 12 Complex 1 125 11 15 250 13 15 500 14 20 Complex 2 125 16 13 250 16 15 500 18 16 DMSO (AR) — — Table 4. Antibacterial activities of the ligands and the complexesThe two Ni(Ⅱ) complexes have moderate to strong antibacterial activity against S. aureus and E. coli and exhibit higher activity than the corresponding Schiff bases. It is interesting to note that 1 has stronger activities against E. coli than 2 whereas 2 has stronger activities against S. aureus than 1, which is similar to the corresponding Schiff bases. The result indicates that chlorine and bromine atoms may have a subtle influence on the antibacterial activities.
-
-
[1]
Saghatforoush L A, Chalabian F, Aminkhani A, et al. Eur. J. Med. Chem., 2009, 44(11):4490-4495 doi: 10.1016/j.ejmech.2009.06.015
-
[2]
Hranjec M, Starčević K, Pavelić S K, et al. Eur. J. Med. Chem., 2011, 46(6):2274-2279 doi: 10.1016/j.ejmech.2011.03.008
-
[3]
Creaven B S, Devereux M, Foltyn A, et al. Polyhedron, 2010, 29(2):813-822 doi: 10.1016/j.poly.2009.11.002
-
[4]
张奇龙, 王焕宇, 江峰, 等.无机化学学报, 2016, 32(3):464-468 https://www.researchgate.net/publication/301584071_Syntheses_Crystal_Structures_and_Bacteriostatic_Activities_of_NiII_and_CuII_Complexes_Based_on_Pentaerythrityltetramine_Schiff_BaseZHANG Qi-Long, WANG Huan-Yu, JIANG Feng, et al. Chinese J. Inorg. Chem., 2016, 32(3):464-468 https://www.researchgate.net/publication/301584071_Syntheses_Crystal_Structures_and_Bacteriostatic_Activities_of_NiII_and_CuII_Complexes_Based_on_Pentaerythrityltetramine_Schiff_Base
-
[5]
仇晓阳.无机化学学报, 2014, 30(7):1667-1672 doi: 10.1021/acs.macromol.6b01976QIU Xiao-Yang. Chinese J. Inorg. Chem., 2014, 30(7):1667-1672 doi: 10.1021/acs.macromol.6b01976
-
[6]
Iqbal A, Siddiqui H L, Ashraf C M, et al. Molecules, 2007, 12(2):245-254 doi: 10.3390/12020245
-
[7]
Khatua S, Kang J, Kim K, et al. Eur. J. Inorg. Chem., 2010, 31(11):5018-5026
-
[8]
Maxim C, Pasatoiu T D, Kravtsov V C, et al. Inorg. Chim. Acta, 2008, 361(14/15):3903-3911 http://med.wanfangdata.com.cn/Paper/Detail?id=PeriodicalPaper_JJ025615397
-
[9]
Dhahagani K, Kumar S M, Chakkaravarthi G, et al. Spectro-chim. Acta Part A, 2014, 117:87-94 doi: 10.1016/j.saa.2013.07.101
-
[10]
Shi L, Ge H M, Tan S H, et al. Eur. J. Med. Chem., 2007, 42(4):558-564 doi: 10.1016/j.ejmech.2006.11.010
-
[11]
Creaven B S, Devereux M, Karcz D, et al. J. Inorg. Biochem., 2009, 103(9):1196-1203 doi: 10.1016/j.jinorgbio.2009.05.017
-
[12]
Ates-Alagoz Z, Yildiz S, Buyukbingol E. Chemotherapy, 2007, 53(2):110-113 doi: 10.1159/000100011
-
[13]
Göker H, Kus C, Boykin D W, et al. Bioorg. Med. Chem., 2002, 10(8):2589-2596 doi: 10.1016/S0968-0896(02)00103-7
-
[14]
Hranjec M, Pavlović G, Marjanović M, et al. Eur. J. Med. Chem., 2010, 45(6):2405-2417 doi: 10.1016/j.ejmech.2010.02.022
-
[15]
Starčević K, Kralj M, Ester K, et al. Bioorg. Med. Chem., 2007, 15(13):4419-4426 doi: 10.1016/j.bmc.2007.04.032
-
[16]
Qiu X H, Tong X L. Acta Crystallogr. Sect. E, 2005, E61:m1470-m1471
-
[17]
Qiu X H, Tong X L. Acta Crystallogr. Sect. E, 2005, E61:m2302-m2304
-
[18]
Qiu X H, Tong X L. Acta Crystallogr. Sect. E, 2006, E62:m841-m842
-
[19]
Qiu X H, Tong X L. Acta Crystallogr. Sect. E, 2006, E62:m977-m979
-
[20]
邱晓航, 赵君, 仝小兰.结构化学, 2006, 25(11):1343-1346 http://www.cnki.com.cn/Article/CJFDTotal-JGHX200609013.htmQIU Xiao-Hang, ZHAO Jun, TONG Xiao-Lan. Chin. J. Struct. Chem., 2006, 25(11):1343-1346 http://www.cnki.com.cn/Article/CJFDTotal-JGHX200609013.htm
-
[21]
Maurya M R, Kumar A, Ebel M, et al. Inorg. Chem., 2006, 45(13):5924-5937 http://www.ncbi.nlm.nih.gov/pubmed/16841997
-
[22]
Maurya M R, Chandrakar A K, Chand S. J. Mol. Catal. A:Chem., 2007, 263(1/2):227-237 http://www.sciencedirect.com/science/article/pii/S138111690700091X
-
[23]
赵海燕, 邱晓航, 申泮文.无机化学学报, 2010, 26(12):2221-2226 http://en.cnki.com.cn/Article_en/CJFDTOTAL-WJHX201012017.htmZHAO Hai-Yan, QIU Xiao-Hang, SHEN Pan-Wen. Chinese J. Inorg. Chem., 2010, 26(12):2221-2226 http://en.cnki.com.cn/Article_en/CJFDTOTAL-WJHX201012017.htm
-
[24]
Kumar R, Kumar R, Mahiya K, et al. Transition Met. Chem., 2015, 40(2):189-195 doi: 10.1007/s11243-014-9905-y
-
[25]
Patil S A, Prabhakara C T, Halasangi B M, et al. Spectrochim. Acta Part A, 2015, 137:641-651 doi: 10.1016/j.saa.2014.08.028
-
[26]
Chai L Q, Zhang H S, Huang J J, et al. Spectrochim. Acta Part A, 2015, 137:661-669 doi: 10.1016/j.saa.2014.08.084
-
[27]
Chandra S, Gautam S, Rajor H K, et al. Spectrochim. Acta Part A, 2015, 137:749-760 doi: 10.1016/j.saa.2014.08.046
-
[28]
Cescon L A, Day A R. J. Org. Chem., 1962, 27(2):581-586 doi: 10.1021/jo01049a056
-
[29]
Sheldrick G M. SADABS, Program for Empirical Absorption Correction of Area Detector Data, University of Göttingen, Germany, 1996.
-
[30]
Sheldrick G M. SHELXL-97, Program for X-ray Crystal Structure Solution, Göttingen University, Germany, 1997.
-
[31]
Drew W L, Barry A L, O'Toole R, et al. Appl. Environ. Microbiol., 1972, 24(2):240-247 http://www.ncbi.nlm.nih.gov/pubmed/5071651
-
[32]
Zhao H Y, Ma J J, Han Z G, et al. Synth. React. Inorg. Met.-Org. Chem., 2015, 45(4):621-627 doi: 10.1080/15533174.2013.841229
-
[33]
Mohapatra S C, Tehlan S, Hundal M S, et al. Inorg. Chim. Acta, 2008, 361(7):1897-1907 doi: 10.1016/j.ica.2007.10.002
-
[34]
Golcu A, Tumer M, Demirelli H, et al. Inorg. Chim. Acta, 2005, 358(6):1785-1797 doi: 10.1016/j.ica.2004.11.026
-
[35]
Demir S, Yazclar T K, Tas M. Inorg. Chim. Acta, 2014, 409(Part A):399-406
-
[1]
-
Figure 2 Formation of 2D network by hydrogen bonding interactions in 1 (a) and 2 (b)
Symmetry codes: ⅱ-x+3/2, y-1/2, -z+3/2; ⅲ-x+1, y, -z+3/2; ⅳ x+1/2, y+1/2, z; ⅴ x+1/2, y-1/2, z; ⅵ x-1/2, y-1/2, z; ⅶ x-1/2, y+1/2, z in (a); ⅱ x, y-1, z; ⅲ x+1/2, y+1/2, z; ⅳ-x+3/2, y+1/2, -z+1/2, ⅴ x+1/2, y+1/2, z; ⅵ x+1/2, y-1/2, z; ⅶ x-1/2, y, -1/2 z; ⅷ x-1/2, y+1/2, z in (b)
Table 1. Crystallographic data and refinement summary for the 1 and 2

Table 2. Selected bond lengths (nm) and bond angles (°) for 1 and 2
1 Ni1-N1 0.208 6(3) Ni1-N1ⅰ 0.208 6(3) Ni1-N2 0.208 2(3) Ni1-N2ⅰ 0.208 2(3) Ni1-O1 0.203 6(2) Ni1-O1ⅰ 0.203 6(2) O1ⅰ-Ni1-O1 94.36(13) N2ⅰ-Ni1-N1ⅰ 88.61(10) O1ⅰ-Ni1-N2ⅰ 174.77(10) N2-Ni1-N1ⅰ 97.13(10) O1-Ni1-N2ⅰ 87.75(10) O1ⅰ-Ni1-N1 87.77(10) O1ⅰ-Ni1-N2 87.75(10) O1-Ni1-N1 86.69(10) O1-Ni1-N2 174.77(10) N2ⅰ-Ni1-N1 97.13(10) N2ⅰ-Ni1-N2 90.55(15) N2-Ni1-N1 88.61(10) O1ⅰ-Ni1-N1ⅰ 86.69(10) N1ⅰ-Ni1-N1 171.85(15) O1-Ni1-N1ⅰ 87.77(10) 2 Ni1-N1 0.208 0(4) Ni1-N1ⅰ 0.208 0(4) Ni1-N2 0.208 9(4) Ni1-N2ⅰ 0.208 9(4) Ni1-O1 0.203 7(3) Ni1-O1ⅰ 0.203 7(3) O1ⅰ-Ni1-O1 94.79(18) N1ⅰ-Ni1-N2ⅰ 88.88(14) O1ⅰ-Ni1-N1ⅰ 86.57(13) N1-Ni1-N2ⅰ 97.56(10) O1-Ni1-N1ⅰ 87.23(13) O1ⅰ-Ni1-N2 87.43(13) O1ⅰ-Ni1-N1 87.23(13) O1-Ni1-N2 174.84(14) O1-Ni1-N1 86.57(13) N1ⅰ-Ni1-N2 97.56(14) N1ⅰ-Ni1-N1 170.8(2) N1-Ni1-N2 88.88(14) O1ⅰ-Ni1-N2ⅰ 174.84(14) N2ⅰ-Ni1-N2 90.7(2) O1-Ni1-N2ⅰ 87.43(13) Symmetry transformations used to generate equivalent atoms: ⅰ-x+1, y, -z+3/2 for 1; ⅰ-x+1, y, -z+1/2 for 2 Table 3. Hydrogen bond parameters in 1 and 2
D-H…A d(D-H)/nm d(H…A)/nm d(D…A)/nm ∠DHA/(°) 1 N3-H3 …02ⅱ 0.086 0.185 7 0.270 4 168.11 02-H2E …N2 0.085 0.248 5 0.322 6 146.28 02-H2F …01ⅲ 0.085 0.191 2 0.264 8 144.09 C1-H1B …C11ⅷ 0.097 0.292 4 0.379 6 150.00 2 N3-H3 …02ⅱ 0.086 0.185 0 0.269 9 169.07 02-H2E …01ⅲ 0.085 0.191 9 0.265 3 143.87 02-H2F …N2ⅳ 0.085 0.249 0 0.323 2 146.31 Symmetry codes: ⅱ-x+3/2, y-1/2, -z+3/2; ⅲ-x+1, y, -z+3/2; ⅷ-x+1, -y+1, -z+1 for 1; ⅱ x, y-1, z; ⅲ x+1/2, y+1/2, z; ⅳ-x+3/2, y+1/2, -z+1/2 for 2 Table 4. Antibacterial activities of the ligands and the complexes
Compound Dose/(μg·mL-1) Inh1b1t1on zone diameter/mm S. aureus E. coli HL1 125 9 13 250 11 14 500 12 16 HL2 125 10 11 250 12 12 500 13 12 Complex 1 125 11 15 250 13 15 500 14 20 Complex 2 125 16 13 250 16 15 500 18 16 DMSO (AR) — — -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 3
- 文章访问数: 808
- HTML全文浏览量: 97


 下载:
下载:



 下载:
下载:

