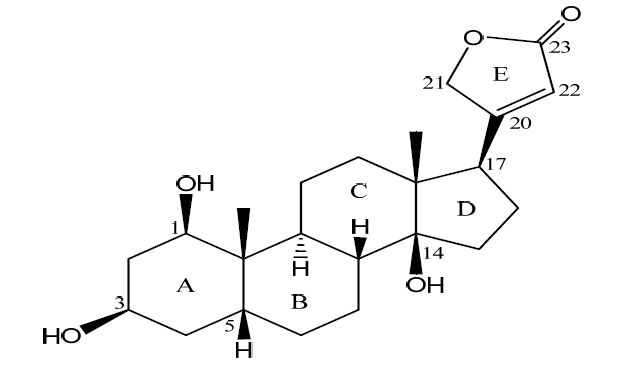
 图Scheme1
Structure of compound 1
Scheme1.
Structure of compound 1
图Scheme1
Structure of compound 1
Scheme1.
Structure of compound 1

Isolation, Crystal Structure and Na+/K+-ATPase Inhibitory Activity of 1β-Hydroxydigitoxigenin①
English
Isolation, Crystal Structure and Na+/K+-ATPase Inhibitory Activity of 1β-Hydroxydigitoxigenin①
-
Key words:
- crystal structure
- / 1β-hydroxydigitoxigenin
- / Na+/K+-ATPase
-
1 INTRODUCTION
Cardiotonic Steroids (CTS) are of significant therapeutic value for the treatment of congestive heart failure and arrhythmia through inhibition of the pumping function of Na+/K+-ATPase[1]. The natural CTS include cardenolides and bufadienolides[2]. Currently, the clinically available CTS-type of drugs, such as digoxin and digitoxin, belong to cardenolides which possess a steroid skeleton bearing a butenolide ring at position C17β and one or more sugar groups at C3[3]. However, this kind of drugs often has narrow therapeutic windows[4]. Thus there is sustained effort to search for structural diverse CTS with significant inhibition on Na+/K+-ATPase but less side effects.
We have been engaged in the identification of structurally unique CTS from animals[5-8] and medicinal plants[9, 10], structural modifications[11, 12], assessment of their effects on Na+/K+-ATPase[7, 10-13], and investigation of the in depth signal transduction pathway[14-16]. Our phytochemical study on the chemical constituents of Streptocaulon Juventas led to the isolation of 1β-hydroxydigitoxigenin (1, Scheme 1), which was found to inhibit the Na+/K+-ATPase in a dose-dependent manner. Insight into the biological and physicochemical functions of CTS with complex stereochemistry at the molecular level requires a precise understanding of their three-dimensional structures. We report herein the three-dimensional structure of compound 1, comparison of con formations between crystalline and solution states and the Na+/K+-ATPase inhibitory activity.
2 EXPERIMENTAL
2.1 Materials and instrumentations
UV spectrum was recorded on a Shimadzu UV- 2450 spectrophotometer. IR spectrum was recorded on a Jasco FT/IR-480 plus spectrophotometer. NMR spectra were measured on a Bruker AM-600 spectrometer at 25 ℃. A Shimadzu LC-20 AT equipped with a SPD-M20A PDA detector was used for HPLC. Thin-layer chromatography (TLC) analyses were carried out using pre-coated silica gel GF254 plates (Qingdao Marine Chemical Plant, Qingdao, People’s Republic of China). Column chromatographies were performed on MCI gel (CHP20P, 75 ~ 50 μm, Mitsubishi Chemical Industries Ltd.) and Sephadex LH-20 gel (Amersham Biosciences) was used for column chromatography. All solvents used were of analytical grade (Guangzhou Chemical Reagents Company, Ltd.).
2.2 Extraction and isolation
The roots of Streptocaulon juventas (20 kg) were chopped into small pieces (0.5 cm long) and extracted with ethanol (2 h, 2×80 L) under reflux conditions. The ethanol extract was evaporated under vacuum to obtain the crude extract (1880 g), which was then suspended in distilled water and partitioned successively with petroleum ether (PE), dichloromethane (CH2Cl2) and ethyl acetate (EtOAc). The EtOAc fraction (80 g) was uploaded on MCI resin column and eluted with a gradient of MeOH-H2O (0:100-100:0, v/v) to afford nineteen fractions (Fr. 1 ~ 9). Fr. 9 (3.5 g, eluted with 60% MeOH) was further purified by Sephadex LH-20 column chromatography, which was eluted with MeOH to afford compound 1 (17.0 mg).
2.3 Spectral structure determination
1β-hydroxydigitoxigenin (1): HRESI-MS (m/z): 391.2000 ([M+H]+); UV (MeOH) λmax 217 nm; IR (KBr): 3570, 1746 cm-1. 1H NMR (CD3OD, 600 MHz) δ: 3.78 [s, H(1)], 4.15 [s, H(3)], 2.85 [s, H(17)], 0.91 [s, H3(18)], 1.11 [s, H3(19)], 5.05 [d, J = 18.6 Hz, H(21α)], 4.93 [d, J = 18.6 Hz H(21β)], 5.92, [s, H(22)]; 13C NMR (CD3OD, 150 MHz) δ: 74.9 C(1), 33.2 C(2), 69.6 C(3), 34.5 C(4), 32.0 C(5), 27.5 C(6), 22.4 C(7), 42.9 C(8), 38.7 C(9), 40.9 C(10), 22.4 C(11), 41.4 C(12), 51.0 C(13), 86.4 C(14), 33.5 C(15), 28.2 C(16), 52.2 C(17), 16.5 C(18), 19.5 C(19), 177.4 C(20), 75.5 C(21), 118.0 C(22), 178.5 C(23).
2.4 X-ray structure determination
The crystals suitable for X-ray structure determination were obtained by slow evaporation of methanol solution at room temperature. A colorless bladelike crystal of the title compound with dimensions of 0.14mm× 0.04mm × 0.01mm was selected and mounted on a thin glass fiber. Due to the extremely small size of the crystal, direct collection of the diffraction data by in house X-ray diffractometer was not successful.
Intensity data were collected at 150 K on a D8 diffractometer equipped with a Bruker PHOTON100 CMOS detector at Beamline 11.3.1 at the Advanced Light Source (Lawrence Berkeley National Laboratory) using synchrotron radiation tuned to λ = 1.2399 Å. For data collection, the frames were measured for a duration of 1 s at 0.5o interval of ω with a maximum 2θ value of ~102o. The data frames were collected using the program APEX2 and processed using the program SAINT routine within APEX2. The data were corrected for absorption and beam corrections based on the multi-scan technique as implemented in SADABS.
A total of 12727 reflections were collected in the range of 3.4<θ<50.99° (index ranges: -9≤h≤9, -16≤k≤16, -12≤l≤12) by using an ω scan mode. Among the 4247 independent reflections (Rint = 0.0509), 3687 reflections with I > 2σ(I) were considered as observed and used in the succeeding refinements. Corrections for incident and diffracted beam absorption effects were applied using SADABS. The structure was solved by direct methods with SHELXS-97 and expanded by using Fourier difference techniques. The non-hydrogen atoms were refined anisotropically, and the hydrogen atoms were added according to theoretical models. The structure was refined by full-matrix leastsquares techniques on F2 with SHELXL-97. The final refinement gave R = 0.0625, wR = 0.1687 (I > 4σ(I), w = 1/[σ2(Fo 2) + (0.0931P)2 + 0.6175P], where P = (Fo 2 + 2Fc 2)/3). (Δ/σ)max<0.001, S = 1.064, (Δρ)max = 0.417, and (Δρ)min = -0.485 e/Å3.
2.5 Na+/K+-ATPase inhibitory activity
The Na+/K+-ATPase inhibitory activities were determined according to the known colormetric method[13]. The purified Na+/K+-ATPase was prepared from pig kidney as described previously[13]. The specific activities of Na+/K+-ATPase of various kidney preparations were in the range of 900~1200 μmol Pi /mg/h, which was more than 95% of the total ATPase activity. The inhibitory activities of different concentrations of compound 1 (purity over 98% by HPLC analysis) on 0.5 μg of Na+/K+- ATPase were determined by measuring the release of inorganic phosphate from ATP over a 15 min period at 37 ℃ in an assay medium containing 100 mM NaCl, 20 mM KCl, 1 mM MgCl2, 1 mM EGTA, 20 mM Tris-HCl and 2 mM ATP-Mg2+ mixture, pH 7.4 buffered to pH 7.40. The absorbance at 620 nm was measured by a microplate reader (SpectraMax 340PC384, Molecular Devices, USA) after colorized by using the Biomol Green Reagent. The GraphPad Prism software (Version 5) was used to fit sigmoid curves to estimate the slopes and IC50 for the dose/inhibition relationships.
3 RESULTS AND DISCUSSION
Compound 1 was obtained by silica gel column chromatography of the 95% ethanol extract followed by open MCI resin column chromatography, and re-crystallized as colorless blade-like crystals from methanol solution. High resolution ESIMS analysis of 1 showed a quasi-molecular ion peak at [M+H]+ 391.2000, corresponding to a molecular formula C23H24O5. The IR spectrum indicated the presence of hydroxy (3570 cm-1) and α, β-unsaturated-γ-lactone (1746 cm-1) groups.
The 1H-NMR data of 1 showed characteristic signals on the lactone ring δ5.05 [d, J = 18.6 Hz, H(21α)], 4.93 [d, J = 18.6 Hz H(21β)] and 5.92 [s, H(22)], two hydroxylated methines δ3.78 [br.s, H(1)] and 4.15 [br.s, H(3)] and two quaternary methyls δ0.91 [s, H3(18)] and 1.11 [s, H3(19)]. These spectroscopic data indicated that compound 1 is a typical cardenolide[10].
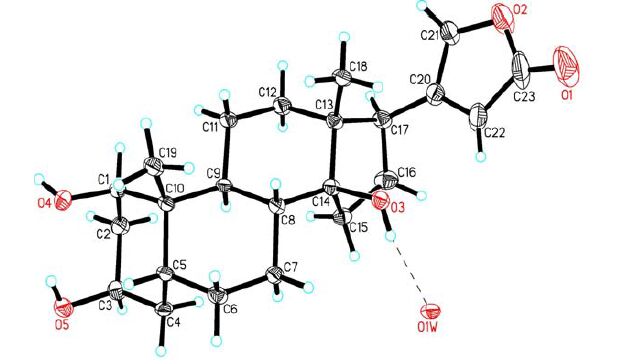
The single crystals of the title compound were obtained by slow evaporation of its methanol solution. To establish the three-dimensional structure of the title compound, a single crystal of 1 was used for X-ray diffraction analysis. X-ray analysis revealed that the crystal belongs to monoclinic system with space group P21, and the asymmetric unit contains a molecule of 1 and a water molecule. Selected bond lengths and bond angles of 1 are given in Table 1. Fig. 1 shows the molecular structure of the title compound, and Fig. 2 depicts the packing diagram.
Bond Dist. Bond Dist. O(1)-C(23) 1.230(8) C(8)-C(9) 1.551(4) O(2)-C(23) 1.364(7) C(9)-C(11) 1.536(4) O(2)-C(21) 1.446(6) C(9)-C(10) 1.569(5) O(3)-C(14) 1.454(4) C(10)-C(19) 1.542(5) O(4)-C(1) 1.446(4) C(11)-C(12) 1.523(6) O(5)-C(3) 1.440(5) C(12)-C(13) 1.544(5) C(1)-C(2) 1.521(6) C(13)-C(18) 1.529(5) C(1)-C(10) 1.553(5) C(13)-C(14) 1.545(4) C(2)-C(3) 1.534(5) C(13)-C(17) 1.581(5) C(3)-C(4) 1.522(5) C(14)-C(15) 1.533(5) C(4)-C(5) 1.541(4) C(15)-C(16) 1.549(5) C(5)-C(6) 1.525(5) C(16)-C(17) 1.553(5) C(5)-C(10) 1.561(4) C(17)-C(20) 1.498(5) C(6)-C(7) 1.535(5) C(20)-C(22) 1.335(6) C(7)-C(8) 1.544(4) C(20)-C(21) 1.495(6) C(8)-C(14) 1.537(5) C(22)-C(23) 1.473(9) Angle (°) Angle (°) C(23)-O(2)-C(21) 107.2(4) C(11)-C(12)-C(13) 114.1(3) O(4)-C(1)-C(2) 108.2(3) C(18)-C(13)-C(12) 109.3(3) O(4)-C(1)-C(10) 108.4(3) C(18)-C(13)-C(14) 112.6(3) C(2)-C(1)-C(10) 114.2(3) C(12)-C(13)-C(14) 108.3(3) C(1)-C(2)-C(3) 111.2(3) C(18)-C(13)-C(17) 114.5(3) O(5)-C(3)-C(4) 111.2(3) C(12)-C(13)-C(17) 107.8(3) O(5)-C(3)-C(2) 110.1(3) C(14)-C(13)-C(17) 104.1(3) C(4)-C(3)-C(2) 109.3(3) O(3)-C(14)-C(15) 107.0(3) C(3)-C(4)-C(5) 113.4(3) O(3)-C(14)-C(8) 108.7(2) C(6)-C(5)-C(4) 110.8(3) C(15)-C(14)-C(8) 116.6(3) C(6)-C(5)-C(10) 111.3(3) O(3)-C(14)-C(13) 106.9(2) C(4)-C(5)-C(10) 112.4(3) C(15)-C(14)-C(13) 103.1(3) C(5)-C(6)-C(7) 112.2(3) C(8)-C(14)-C(13) 114.0(3) C(6)-C(7)-C(8) 110.9(3) C(14)-C(15)-C(16) 103.8(3) C(14)-C(8)-C(7) 111.9(3) C(15)-C(16)-C(17) 106.8(3) C(14)-C(8)-C(9) 113.0(2) C(20)-C(17)-C(16) 113.5(3) C(7)-C(8)-C(9) 111.3(2) C(20)-C(17)-C(13) 114.4(3) C(11)-C(9)-C(8) 109.8(3) C(16)-C(17)-C(13) 105.0(3) C(11)-C(9)-C(10) 113.1(3) C(22)-C(20)-C(21) 106.9(4) C(8)-C(9)-C(10) 112.0(3) C(22)-C(20)-C(17) 132.2(4) C(19)-C(10)-C(1) 107.2(3) C(21)-C(20)-C(17) 120.8(3) C(19)-C(10)-C(5) 109.4(3) O(2)-C(21)-C(20) 107.1(4) C(1)-C(10)-C(5) 107.9(3) C(20)-C(22)-C(23) 109.5(5) C(19)-C(10)-C(9) 111.3(3) O(1)-C(23)-O(2) 118.1(6) C(1)-C(10)-C(9) 111.2(3) O(1)-C(23)-C(22) 132.3(6) C(5)-C(10)-C(9) 109.8(3) O(2)-C(23)-C(22) 109.1(5) C(12)-C(11)-C(9) 112.1(3) 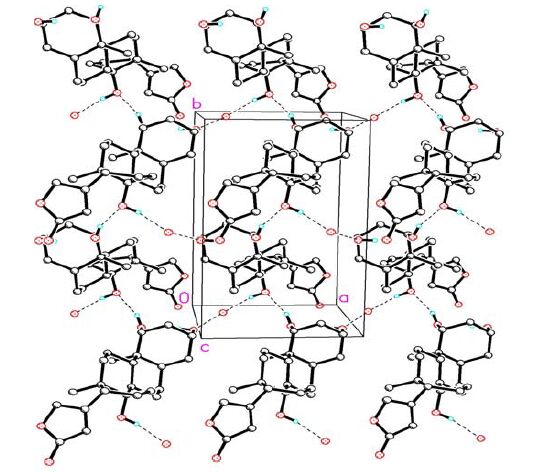 图2
A packing diagram for compound 1. Hydrogen-bonding network of 1 viewed roughly down the c-axis. Selected hydrogen atoms highlight the scheme of the hydrogen bonding
Figure2.
A packing diagram for compound 1. Hydrogen-bonding network of 1 viewed roughly down the c-axis. Selected hydrogen atoms highlight the scheme of the hydrogen bonding
图2
A packing diagram for compound 1. Hydrogen-bonding network of 1 viewed roughly down the c-axis. Selected hydrogen atoms highlight the scheme of the hydrogen bonding
Figure2.
A packing diagram for compound 1. Hydrogen-bonding network of 1 viewed roughly down the c-axis. Selected hydrogen atoms highlight the scheme of the hydrogen bonding
D-H…A d(D-H) d(H-A) d(D…A) ZDHA O(3)—H(3)…O(1w) a 0.82 2.573 2.713 (4) 166.8 O(4)-H(4)…O(3)b 0.82 2.139 2.685 (3) 160.5 O(1w)—H(1w)-O(5)c 0.82 - 2.705 - Symmetry codes: (a) x, y, z; (b) 1–x, 0.5+y, 1–z; (c) 2–x, y–0.5, 1–z; (-) mean not determined due to the absence of hydrogen on the water molecule Compound 1 is composed of three cyclohexane rings (A, B and C), one saturated five-membered ring (D) and one lactone ring (E). The stereochemistry of the ring juncture is A/B cis, B/C trans. Three hydroxy groups were located at the C(1β), C(3β) and C(14β) positions of the steroidal skeleton.
The cyclohexane rings A, B and C exist in normal chair conformations. The five-membered ring D adopts an envelope conformation with C(14) displaced by 0.6145(2) from the mean plane of the remaining four atoms [C(13), C(15), C(16) and C(17) with a mean deviation of 0.0093 Å]. The lactone ring E is planar with a mean deviation of 0.0123 Å, and makes a dihedral angle of 78.7° with the envelope plane of ring D, which is similar to the corresponding angle in Δ14, 15-anhydro-24-thiocarbonylbufalin[12], a bufadienolide derivative modified from bufalin. Due to the absence of heavy atom in the molecule, the final refinement resulted in a non-significant Flack parameter 0.1(5); however, considering the conserved C(17β)-orientated lactone ring, the absolute configuration of compound 1 could be assigned, as shown in Fig. 1.
In solid state, the water molecule was linked into compound 1 through hydrogen bond O(3)-H(3)… O(1w) (2.713 Å, ). Compound 1 is further linked into a chain through hydrogen bond O(4)-H(4)…O(3) (2.685 Å, symmetry code: 1-x, 0.5+y, 1-z). Two adjacent chains were inter- connected through hydrogen bond O(1w)-H(1w)…O(5) (2.705 Å, symmetry code: 2-x, y-0.5, 1-z) into a network (Fig. 2).
Normally, molecules exert biological functions in solution state. Thus, it is necessary to compare the solution conformation with the solid state conformation. The broad single peak of H(3) indicated that the coupling between H(2α, 2β) and H(3), H(3) and H(4α, 4β) is small. X-ray analysis showed that the H(3) is on the equatorial position (e) and the hydroxyl group on the axial position (a). Consequently, H(3) makes ee- and ea-type small couplings with H(2α, 2β) and H(4α, 4β), resulting in a broad single peak. Similarly, broad single peak of H(1) in the 1H-NMR spectrum is also consistent with the β-orientated 1-OH revealed by X-ray analysis. In addition, 1H-NMR spectrum of 1 showed that the coupling constant between H(17) and H(16α) is 5.4 Hz, which is consistent with the large torsion angle H(17)-C(17)-C(16)-H(16α) 62.5°. Similarly, the coupling constant between H(17) and H(16β) is 9.0 Hz which is consistent with small torsion angle H(17)-C(17)-C(16)-H(16β) 7.5°. Thus the confortion of compound 1 in crystalline state is consistent with the solution structure in methanol, which is similar to cinobufagin 3-hemisuberate methyl ester[8] and thiocarbonylbufalin[12].
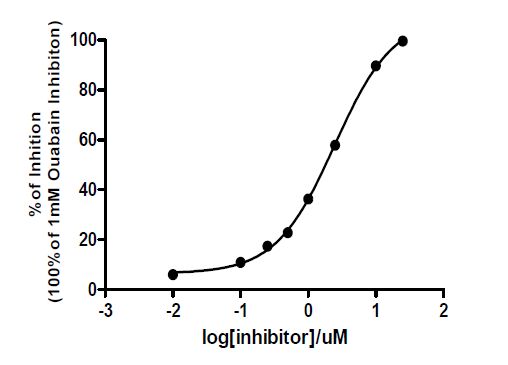
The inhibition on Na+/K+-ATPase property of 1 was studied using an enzymatic assay. Compound 1 showed significant inhibition on Na+/K+- ATPase with IC50 value of 2.46 μM in a dose-dependent manner (Fig. 3), which is stronger than thiocarbonylbufalin[12] but weaker than a close analog digitoxigenin (IC50 = 0.132 μM)[17], indicating that a lactone ring is important and the substitution of a hydroxyl group at C(1) is not favor for the inhibition of Na+/K+-ATPase.
ACKNOWLEDGEMENTS
The Advanced Light Source for diffraction data collection is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DEAC02- 05CH11231. The first author would like to thank Marshall Institute for Interdisciplinary Research for providing scholarship to perform the research work in Marshall University.
-
-
[1]
Skou J. C.. The identification of the sodium-potassium pump (Nobel Lecture). Angew[J]. Chem. Int. Ed., 1998, 37: 2320-2328. doi: 10.1002/(ISSN)1521-3773
-
[2]
Newman R. A., Yang P., Pawlus A. D., Block K. I.. Cardiac glycosides as novel cancer therapeutic agents[J]. Mol. Interv., 2008, 8: 36-49. doi: 10.1124/mi.8.1.8
-
[3]
Prassas I., Diamandis E. P.. Novel therapeutic applications of cardiac glycosides[J]. Nat. Rev. Drug Discov., 2008, 7: 926-935. doi: 10.1038/nrd2682
-
[4]
Barrueto F., Kirrane B. M., Cotter B. W., Hoffman R. S., Nelson L. S.. Cardioactive steroid poisoning: a comparison of plant- and animal-derived compounds[J]. J. Med. Toxicol., 2006, 2: 152-155. doi: 10.1007/BF03161183
-
[5]
Tian H. Y., Wang L., Zhang X. Q., Zhang D. M., Wang Y., Jiang R. W., Ye W. C.. New bufadienolides and C23 steroids from the venom of Bufo bufo gargarizans[J]. Steroids, 2010, 75: 884-890. doi: 10.1016/j.steroids.2010.05.013
-
[6]
Tian H. Y., Wang L., Zhang X. Q., Wang Y., Zhang D. M., Jiang R.W., Liu Z., Liu J. S., Li Y. L., Ye W. C.. Bufogargarizins A and B, two novel 19-norbufadienolides with unprecedented skeletons from the venom of Bufo bufo gargarizans[J]. Chem. Eur. J., 2010, 16: 10989-10993. doi: 10.1002/chem.v16:36
-
[7]
Tian H. Y., Luo S. L., Liu J. S., Wang L., Wang Y., Zhang D. M., Zhang X. Q., Jiang R. W., Ye W. C.. C23 Steroids from the venom of Bufo bufo gargarizans[J]. J. Nat. Prod., 2013, 76: 1842-1847. doi: 10.1021/np400174f
-
[8]
Zhang Y., Feng J., Ye W. C., Tian H. Y., Jiang R. W.. Crystal structure and anticancer properties of cinobufagin 3-hemisuberate methyl ester[J]. Chin. J. Struct. Chem., 2014, 33: 790-794.
-
[9]
Cheng W., Tan Y. F., Tian H. Y., Gong X. W., Chen K. L., Jiang R.W.. Two new bufadienolides from the rhizomes of Helleborus thibetanus with inhibitory activities against prostate cancer cells[J]. Nat. Prod. Res., 2014, 28: 901-908. doi: 10.1080/14786419.2014.891200
-
[10]
Zhang R. R., Tian H. Y., Tan Y. F., Chung T. Y., Sun X. H., Xia X., Ye W. C., Middleton D. A., Fedosova N., Esmann M., Tzen J. T. C., Jiang R. W.. Structures, chemotaxonomic significance, cytotoxic and Na+,K+-ATPase inhibitory activities of new cardenolides from Asclepias curassavica[J]. Org. Biomol. Chem., 2014, 12: 8919-8929. doi: 10.1039/C4OB01545B
-
[11]
Tian H. Y., Yuan X. F., Jin L., Li J., Luo C., Ye W. C., Jiang R.W.. A bufadienolide derived androgen receptor antagonist with inhibitory activities against prostate cancer cells[J]. Chem. Biol. Interact., 2014, 207: 16-22. doi: 10.1016/j.cbi.2013.10.020
-
[12]
Zhang P. W., Tang H. J., Tian H. Y., Zhang R. R., Jiang R. W.. Synthesis, crystal structure and Na+/K+-ATPase inhibitory activity of Δ4,15-anhydro-24-thiocarbonylbufalin[J]. Chin. J. Struct. Chem., 2014, 8: 1123-1128.
-
[13]
Zhang Z., Li Z., Tian J., Jiang W., Wang Y., Zhang X., Li Z., You Q. D., Shapiro J. I., Si S., Xie Z.. Identification of hydroxyxanthones as Na/K-ATPase ligands[J]. Mol. Pharmacol., 2010, 77: 961-967. doi: 10.1124/mol.110.063974
-
[14]
Xie Z.. Molecular mechanisms of Na/K-ATPase-mediated signal transduction[J]. Ann. N.Y. Acad. Sci., 2003, 986: 497-503. doi: 10.1111/nyas.2003.986.issue-1
-
[15]
Pierre S. V., Yang C., Yuan Z., Seminerio J., Mouas C., Garlid K. D., Dos-Santos P., Xie Z.. Ouabain triggers preconditioning through activation of the Na+,K+-ATPase signaling cascade in rat hearts[J]. Cardiovasc. Res., 2007, 73: 488-496. doi: 10.1016/j.cardiores.2006.11.003
-
[16]
Lai F., Madan N., Ye Q., Duan Q., Li Z., Wang S., Si S., Xie Z.. Identification of a mutant α1 Na/K-ATPase that pumps but is defective in signal transduction[J]. J. Biol. Chem., 2013, 288: 13295-13304. doi: 10.1074/jbc.M113.467381
-
[17]
Cornelius F., Kanai R., Toyoshima C.. A structural view on the functional importance of the sugar moiety and steroid hydroxyls of cardiotonic steroids in binding to Na,K-ATPase[J]. J. Biol. Chem., 2013, 288: 6602-6616. doi: 10.1074/jbc.M112.442137
-
[1]
-
Table 1. Selected Bond Lengths (Å) and Bond Angles (º)
Bond Dist. Bond Dist. O(1)-C(23) 1.230(8) C(8)-C(9) 1.551(4) O(2)-C(23) 1.364(7) C(9)-C(11) 1.536(4) O(2)-C(21) 1.446(6) C(9)-C(10) 1.569(5) O(3)-C(14) 1.454(4) C(10)-C(19) 1.542(5) O(4)-C(1) 1.446(4) C(11)-C(12) 1.523(6) O(5)-C(3) 1.440(5) C(12)-C(13) 1.544(5) C(1)-C(2) 1.521(6) C(13)-C(18) 1.529(5) C(1)-C(10) 1.553(5) C(13)-C(14) 1.545(4) C(2)-C(3) 1.534(5) C(13)-C(17) 1.581(5) C(3)-C(4) 1.522(5) C(14)-C(15) 1.533(5) C(4)-C(5) 1.541(4) C(15)-C(16) 1.549(5) C(5)-C(6) 1.525(5) C(16)-C(17) 1.553(5) C(5)-C(10) 1.561(4) C(17)-C(20) 1.498(5) C(6)-C(7) 1.535(5) C(20)-C(22) 1.335(6) C(7)-C(8) 1.544(4) C(20)-C(21) 1.495(6) C(8)-C(14) 1.537(5) C(22)-C(23) 1.473(9) Angle (°) Angle (°) C(23)-O(2)-C(21) 107.2(4) C(11)-C(12)-C(13) 114.1(3) O(4)-C(1)-C(2) 108.2(3) C(18)-C(13)-C(12) 109.3(3) O(4)-C(1)-C(10) 108.4(3) C(18)-C(13)-C(14) 112.6(3) C(2)-C(1)-C(10) 114.2(3) C(12)-C(13)-C(14) 108.3(3) C(1)-C(2)-C(3) 111.2(3) C(18)-C(13)-C(17) 114.5(3) O(5)-C(3)-C(4) 111.2(3) C(12)-C(13)-C(17) 107.8(3) O(5)-C(3)-C(2) 110.1(3) C(14)-C(13)-C(17) 104.1(3) C(4)-C(3)-C(2) 109.3(3) O(3)-C(14)-C(15) 107.0(3) C(3)-C(4)-C(5) 113.4(3) O(3)-C(14)-C(8) 108.7(2) C(6)-C(5)-C(4) 110.8(3) C(15)-C(14)-C(8) 116.6(3) C(6)-C(5)-C(10) 111.3(3) O(3)-C(14)-C(13) 106.9(2) C(4)-C(5)-C(10) 112.4(3) C(15)-C(14)-C(13) 103.1(3) C(5)-C(6)-C(7) 112.2(3) C(8)-C(14)-C(13) 114.0(3) C(6)-C(7)-C(8) 110.9(3) C(14)-C(15)-C(16) 103.8(3) C(14)-C(8)-C(7) 111.9(3) C(15)-C(16)-C(17) 106.8(3) C(14)-C(8)-C(9) 113.0(2) C(20)-C(17)-C(16) 113.5(3) C(7)-C(8)-C(9) 111.3(2) C(20)-C(17)-C(13) 114.4(3) C(11)-C(9)-C(8) 109.8(3) C(16)-C(17)-C(13) 105.0(3) C(11)-C(9)-C(10) 113.1(3) C(22)-C(20)-C(21) 106.9(4) C(8)-C(9)-C(10) 112.0(3) C(22)-C(20)-C(17) 132.2(4) C(19)-C(10)-C(1) 107.2(3) C(21)-C(20)-C(17) 120.8(3) C(19)-C(10)-C(5) 109.4(3) O(2)-C(21)-C(20) 107.1(4) C(1)-C(10)-C(5) 107.9(3) C(20)-C(22)-C(23) 109.5(5) C(19)-C(10)-C(9) 111.3(3) O(1)-C(23)-O(2) 118.1(6) C(1)-C(10)-C(9) 111.2(3) O(1)-C(23)-C(22) 132.3(6) C(5)-C(10)-C(9) 109.8(3) O(2)-C(23)-C(22) 109.1(5) C(12)-C(11)-C(9) 112.1(3) Table 2. Hydrogen Bond Lengths (Å) and Bond Angles (°)
D-H…A d(D-H) d(H-A) d(D…A) ZDHA O(3)—H(3)…O(1w) a 0.82 2.573 2.713 (4) 166.8 O(4)-H(4)…O(3)b 0.82 2.139 2.685 (3) 160.5 O(1w)—H(1w)-O(5)c 0.82 - 2.705 - Symmetry codes: (a) x, y, z; (b) 1–x, 0.5+y, 1–z; (c) 2–x, y–0.5, 1–z; (-) mean not determined due to the absence of hydrogen on the water molecule -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 5
- 文章访问数: 1901
- HTML全文浏览量: 194


 下载:
下载:



 下载:
下载:

