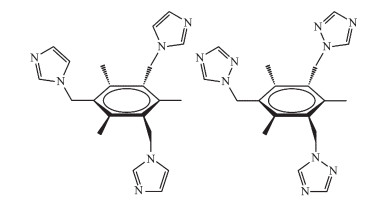
 图Scheme 1
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
Scheme1.
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
图Scheme 1
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
Scheme1.
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB

基于柔性三齿配体的镉(Ⅱ)和锌(Ⅱ)配合物的合成、结构和荧光性质
English
Syntheses, Crystal Structures and Fluorescence of Cadmium(Ⅱ) and Zinc(Ⅱ) Complexes Based on Flexible Tripodal Ligands
-
Key words:
- flexible tripodal ligand
- / coordination polymers
- / luminescence property
-
In recent years, the rational design and assembly of metal organic frameworks (MOFs) have attracted great attention due to their intriguing framework structures and potential applications in optical, gas storage, catalysis, and so on[1-8]. In general, the resulted structures of MOFs could be influenced by several factors, such as the selection of metal centers, organic ligands and the reaction pathways[9]. In contrast to the rigid ligands with fixed conformations, flexible ligands may adopt variable conformations to satisfy the require-ment of the center metal ions[10-11]. In previous work, the utilization of flexible tripodal nitrogen-including ligands have been proved to be effective candidates to build MOF structures, such as 1, 3, 5-tris(4-pyridyl-methyl)benzene[12], 1, 3, 5-tris(pyrazol-1-ylmethyl)-2, 4, 6-triethylbenzene[13], 1, 3, 5-tris(1, 2, 4-triazol-1-ylmethyl)benzene[14], and 1, 3, 5-tris(benzimidazole-1-ylmethyl)-2, 4, 6-trimethylbenzen[15]. Herein, the structures and prop-erties of two two-dimensional (2D) coordination poly-mers based on the 1, 3, 5-tris(imidazol-1-ylmethyl)-2, 4, 6-trimethylbenzene (TITMB) and 1, 3, 5-tris(triazol-1-ylmethyl)-2, 4, 6-trimethylbenzene (TTTMB) have been synthesized and characterized, where both TITMB and TTTMB ligands have adopted cis, cis, trans-configura-tion (Scheme 1)[16-18].
 图Scheme 1
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
Scheme1.
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
图Scheme 1
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
Scheme1.
Schematic showing of the cis, cis, trans-configuration of ligands TITMB and TTTMB
1 Experimental
1.1 Materials and measurements
All commercially available chemicals are of reagent grade and were used as received without further purification. TITMB and TTTMB were prepared according to the literature methods[19]. The C, H and N analyses were performed on a Perkin-Elmer 240C elemental analyzer. Infrared (IR) spectra were recorded on a NEXUS 670 spectrophotometer by using KBr pellets. Thermogravimetric analyses (TGA) were performed on a Perkin Elmer instruments Diamond Thermogravimetric/Differential Thermal Anal-yzer under nitrogen with a heating rate of 10 ℃·min-1. The luminescence spectra were measured at room temperature on PEKIN ELMER Luminescence Spec-trometer LS50B.
1.2 Synthesis of complex {[Cd3(SO4)4(TITMB)2(H2O)4][Cd(H2O)6]·2CH3OH}n (1)
A mixture of CdSO4·8/3H2O (8.54 mg, 0.041 mmol), ligand TITMB (16.6 mg, 0.046 mmol), and 1, 4 -bis(trazol-1-ylmethyl)-2, 3, 5, 6-tetramethylbenzene (p -BTTMB) (12.4 mg, 0.042 mmol) was suspended in 4 mL mixed solvents of methanol and H2O (1:3, V/V), and heated in a Teflon-lined steel bomb at 120 ℃ for 2 d. After the mixture was slowly cooled to room temperature, colorless crystals of 1 were obtained. Anal. Calcd. for C44H76Cd4N12O28S4(%): C, 29.37; H, 4.25; N, 9.34. Found(%): C, 29.28; H, 3.88; N, 9.00. IR (KBr, cm-1): 3 381(s), 1 621(m), 1 520(m), 1 457(w), 1 400(w), 1 319(m), 1 246(m), 1 237(m), 1 118(s), 980(w), 935(w), 760(m), 660(m).
1.3 Synthesis of {[Zn(TTTMB) (HCOO)2]·CH3OH}n (2)
A mixture of Zn(CH3COO)2·2H2O (87.2 mg, 0.4 mmol), TTTMB (145.6 mg, 0.4 mmol), 1, 3-bis(1-imidazo)-1-ylmethyl)-2, 4, 6-trimethylbenzen (m-BITMB) (112.0 mg, 0.4 mmol), 3 mL methanol and 1 mL ethanol was placed in a 10 mL glass vial and dissolved by ultrasound. Methanoic acid was added to adjust pH value to 4. The glass vial was transferred into a 30 mL Teflon-lined stainless steel vessel and heated at 80 ℃ for 3 d. After the mixture was slowly cooled to the room temperature, colorless crystals of 2 were obtained. Anal. Calcd. for C21H27N9O5Zn(%): C, 45.74; H, 4.90; N, 22.87. Found(%): C, 45.00; H, 4.80; N, 22.58. IR (KBr, cm-1): 3 398(m), 3 087(m), 3 005(m), 2 821(m), 2 744(w), 1 712(w), 1 611(s), 1 524(m), 1 448(m), 1 339(s), 1 280(s), 1 221(w), 1 201(w), 1 132(s), 1 033(w), 1 013(w), 1 005(w), 994(s), 875(w), 773(m), 674(s).
1.4 X-ray crystal structure determinations
The single crystals of both complexes 1 and 2 were mounted onto thin glass fibers by epoxy glue in air for data collection. And the diffraction data for complexes 1 and 2 were collected on a Bruker Apex 2 CCD with Mo Kα radiation (λ=0.071 073 nm) using ω-2θ scan method at 298 K. An empirical absorption correction was applied. All the non-hydrogen atoms were refined anisotropically. The hydrogen atoms of organic molecule were refined in calculated positions, assigned isotropic thermal parameters, and allowed to ride their parent atoms, while the H atoms for water are located from different map. All calculations were performed using the SHELX-97 program package[20]. In complex 1, the sulfate oxygen atoms (O1, O2, O4 and O6, O7, O8) split to two parts with the occupation of 0.66(2) and 0.34(2), 0.59(3) and 0.41(3), respe-ctively. Details of the crystal parameters, data collec-tions and refinements for 1 and 2 are summarized in Table 1. Selected bond lengths and angles, and hydrogen bonds for 1 and 2 are listed in Table 2 and Table 3, respectively.
Complex 1 2 Empirical formula C44H76Cd4N12O28S4 C21H27N9O5Zn Formula weight 1 799.01 550.89 Crystal system Trielinic Triclinic Space group P1 P1 a / nm 1.027 61(11) 0.914 93(9) b / nm 1.233 42(15) 1.259 85(13) c / nm 1.469 65(16) 1.291 34(15) α/(°) 68.646(1) 61.093(1) β/(°) 84.642(2) 84.690(2) γ/(°) 69.495(1) 68.873(1) V / nm3 1.623 7(3) 1.209 4(2) Z 1 2 Dc / (g·cm-3) 1.846 1.513 μ/mm-1 1.51 1.07 F(000) 904 572 GOF 1.028 1.047 Collected reflection 8 251 6 403 Unique reflection 5 587 4 186 R1 0.060 0.093 wR2 0.184 0.235 Complex 1 Cd1-N9 0.221 4(7) Cd2-N6 0.225 1(7) Cell-01 0.257(2) Cd1-N3 0.224 0(6) Cd2-05 0.234 3(6) Cel 1-09 0.262 0(10) Cd1-O2C 0.225 0(12) Cd2-O10 0.239 1(8) Cd3-012 0.230 8(5) Cd1-O6B 0.239 7(18) Cd3-O13 0.223 4(8) Cd3-O11 0.232 2(6) Cd1-O2 0.262 8(12) N9-Cd1-N3 173.7(3) O6B-Cd1-O2 162.1(5) N3-Cd1-O6B 80.9(5) N9-Cd1-O2C 94.4(4) O6B-Cd1-O1 142.7(7) N3-Cd1-O2C 91.9(4) N9-Cd1-O2 91.6(3) O9-Cd1-O2 119.7(4) N3-Cd1-O9 84.2(3) N9-Cd1-O6B 97.3(5) O9-Cd1-O1 67.0(7) N3-Cd1-O1 90.2(7) N9-Cd1-O9 89.5(3) O9-Cd1-O6B 76.1(5) N3-Cd1-O2 91.8(3) N9-Cd1-O1 87.7(7) O1-Cd1-O2 52.8(7) O2C-Cd1-O6B 104.3(5) O2C-Cd1-O9 176.0(4) O2C-Cd1-O1 112.2(7) O2C-Cd1-O2 59.4(5) N6-Cd2-O5 89.0(2) O13-Cd3-O12 91.9(3) N6-Cd2-O10 84.6(3) O5D-Cd2-O5 180 O13-Cd3-O12A 88.1(3) N6D-Cd2-N6 180 N6-Cd2-O10D 95.4(3) O12-Cd3-O12A 180 N6-Cd2-O5D 91.0(2) O5-Cd2-O10D 90.2(2) O12-Cd3-O11A 92.9(2) O5-Cd2-O10 89.8(2) O12-Cd3-O11 87.1(2) 013A-Cd3-O11 89.2(3) O10D-Cd2-O10 180 O13A-Cd3-O13 180 O13-Cd3-O11 90.8(3) O11A-Cd3-O11 180 Complex 2 Zn1-O3 0.207 2(6) Zn1-N6 0.207 5(6) Zn1-02 0.217 2(8) Zn1-N3 0.209 3(6) Zn1-N9 0.226 1(6) Zn1-O1 0.238 9(8) O3-Zn1-N6 91.1(2) O3-Zn1-N3 91.9(2) O3-Zn1-O1 92.9(3) N6-Zn1-N3 108.0(2) O3-Zn1-O2 91.6(3) N3-Zn1-O1 99.1(3) N6-Zn1-O2 94.8(3) N3-Zn1-O2 156.9(3) N9-Zn1-O1 83.4(2) O3-Zn1-N9 176.2(2) N6-Zn1-N9 91.9(2) N6-Zn1-O1 152.4(3) N3-Zn1-N9 89.5(2) O2-Zn1-N9 85.8(2) O2-Zn1-O1 57.8(3) Symmetry codes: A: 2-x, -y, -z; B: x, 1+y, z; C: 1-x, 2-y, -z; D: -x+2, -y+2, -z+1 for 1.  Table 3.
Hydrogen bond lengths (nm) and angles (°) for complexes 1~2 Symmetry codes: A: x, -1+y, z; B: 1-x, 1-y, -z for 1.
Table 3.
Hydrogen bond lengths (nm) and angles (°) for complexes 1~2 Symmetry codes: A: x, -1+y, z; B: 1-x, 1-y, -z for 1.
D-H‧‧‧A d(D-H) / nm d(H‧‧‧A) /nm d(D‧‧‧A) / nm ∠D-H‧‧‧A /(°) 1 O9-H9A‧‧‧O5A 0.085 0 0.195 7 0.278 7(14) 165 O9-H9B‧‧‧O14 0.095 0 0.229 1 0.300(2) 141 O10-H10A‧‧‧O8 0.084 9 0.257 6 0.270(2) 123 O11-H11A‧‧‧O7A 0.085 0 0.214 9 0.299 6(13) 174 O11-H11B‧‧‧O4B 0.085 0 0.204 7 0.277 5(19) 143 O12-H12A‧‧‧O3B 0.085 0 0.191 2 0.274 6(12) 167 O12-H12B‧‧‧O6A 0.085 1 0.212 3 0.296(2) 167 O13-H13C‧‧‧O8A 0.085 0 0.207 9 0.271(2) 130 O14-H14‧‧‧O1 0.082 0 0.223 0 0.284(3) 131 2 O5-H5‧‧‧O1 0.081 6 0.196 2 0.276(2) 166 C6-H6‧‧‧O1 0.093 0 0.253 5 0.308 4(12) 118 C2-H2‧‧‧O4 0.093 0 0.217 3 0.309 5(11) 171 Hydrogen atoms and methanol molecules are omitted for clarity; Symmetry codes: A: 2-x, -y, -z; B: x, 1+y, z; C: 1-x, 2-y, -z; D: 2-x, 2-y, 1-z CCDC: 808063, 1; 808067, 2.
2 Results and discussion
2.1 Crystal structure of complex 1
Single crystal X-ray diffraction reveals that complex 1 crystallizes in the triclinic P1 space group. There are two independent units in complex 1: [Cd3(SO4)4(TITMB)2(H2O)4]2- and [Cd(H2O)6]2+. In com-plex 1, three crystallographic distinct Cd(Ⅱ) ions exist (Fig. 1): one seven coordinated Cd1 with bipyramidal geometry and two six coordinated Cd2 and Cd3 with distorted octahedral geometry(Fig. 1a). Each Cd1 is coordinated by two nitrogen atoms (N9B and N3B) from imidazole rings, four oxygen atoms (O1B, O2B, O2C and O6) from three distinct sulfate anions and one terminated water molecule (O9B). Each Cd2 sitting at symmetric center is coordinated by two nitrogen atoms (N6 and N6D) from two imidazole rings, two oxygen atoms (O5 and O5D) from two different sulfate anions and two terminal water molecules (O10 and O10D). Each Cd3 was coordinated by six water molecules and independently located in complex 1. The average bond length of Cd-N and Cd-O is comparable to previously reported[21].
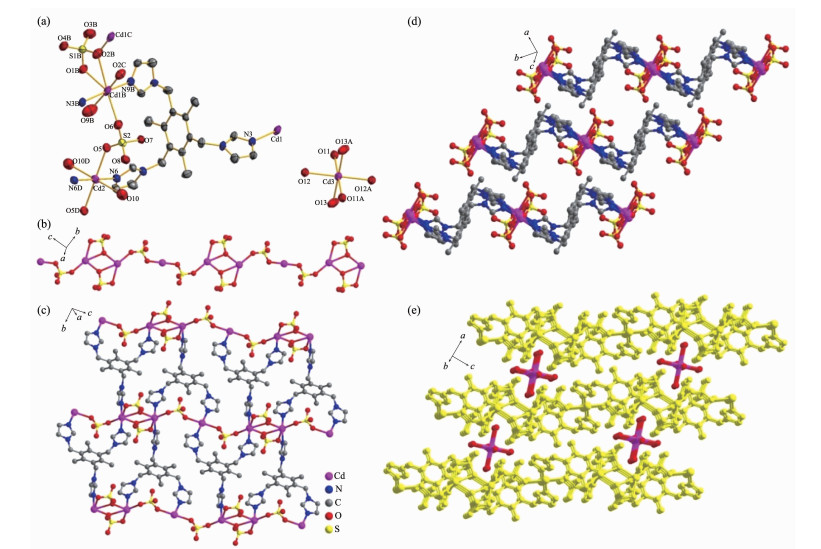 图1
(a) View of ORTEP drawing of complex 1 with 30% ellipsoid; (b) One dimensional inorganic chain formed by bridging sulfate anion and Cd(Ⅱ); (c) Two dimensional network constructed by TITMB ligands with 1D inorganic chain; (d) AAA stacking modes in complex 1; (e) Cd(H2O)6 units located between the 2D networks further extending the 2D network to 3D framework through hydrogen bonds
Figure1.
(a) View of ORTEP drawing of complex 1 with 30% ellipsoid; (b) One dimensional inorganic chain formed by bridging sulfate anion and Cd(Ⅱ); (c) Two dimensional network constructed by TITMB ligands with 1D inorganic chain; (d) AAA stacking modes in complex 1; (e) Cd(H2O)6 units located between the 2D networks further extending the 2D network to 3D framework through hydrogen bonds
图1
(a) View of ORTEP drawing of complex 1 with 30% ellipsoid; (b) One dimensional inorganic chain formed by bridging sulfate anion and Cd(Ⅱ); (c) Two dimensional network constructed by TITMB ligands with 1D inorganic chain; (d) AAA stacking modes in complex 1; (e) Cd(H2O)6 units located between the 2D networks further extending the 2D network to 3D framework through hydrogen bonds
Figure1.
(a) View of ORTEP drawing of complex 1 with 30% ellipsoid; (b) One dimensional inorganic chain formed by bridging sulfate anion and Cd(Ⅱ); (c) Two dimensional network constructed by TITMB ligands with 1D inorganic chain; (d) AAA stacking modes in complex 1; (e) Cd(H2O)6 units located between the 2D networks further extending the 2D network to 3D framework through hydrogen bonds
Each sulfate anion in complex 1 coordinates two Cd(Ⅱ) ions through adopting two different bridging modes (η1-η1-η0-η0-and η1-η2-η0-η0-coordination mode for the four oxygen atoms, respectively), which extends to one dimensional (1D) inorganic chain (Fig. 1b). The 1D chains are further linked by the TITMB ligand with cis, cis, trans-configuration to two dimensional (2D) network (Fig. 1c), where each TITMB ligand coor-dinates to three Cd(Ⅱ)centers. The 2D networks stack in an AAA sequence along the b axis (Fig. 1d), where the [Cd(H2O)6]2+ units locate in the cavity between two adjacent 2D layers by hydrogen bonds to neutralize the charge (Fig. 1e). The strong O-H…O hydrogen bonds between the Cd(H2O)6 units and the sulfate anions facilitated the formation of the 3D structure of complex 1, where the distance between the donor oxygen atom to the acceptor oxygen atom was in the range of 0.270(2)~0.300(2) nm (Table 3).
2.2 Crystal structure of complex 2
As shown in Fig. 2, the asymmetric unit of complex 2 contains one zinc(Ⅱ), one TTTMB ligand and two methanoic acid, and one methanol molecular. Single crystal X-ray crystallographic analyses reveal that each zinc(Ⅱ) is six-coordinated with a distorted octahedral geometry by three nitrogen atoms (N3, N6 and N9) from three different TTTMB ligands and three oxygen atoms (O1, O2, O3) from two methanoic acid (Fig. 2a). Two methanoic acids present two different coordination modes, chelate and monodentate, respec-tively[17]. The Zn-O bond distances vary from 0.207 2(6) to 0.238 9(8) nm, and the Zn-N distances are in the range of 0.207 5(6)~0.226 1(6) nm.
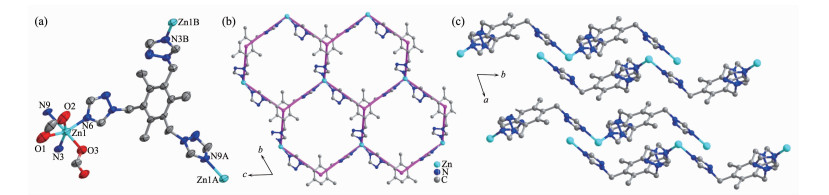 图2
(a) View of ORTEP of complex 2 with 30% ellipsoid; (b) Two dimensional honeycomb 63 network built by TTTMB ligand with Zn(Ⅱ) centers; (c) ABAB packing mode of 2D layers along a axis in complex 2
Figure2.
(a) View of ORTEP of complex 2 with 30% ellipsoid; (b) Two dimensional honeycomb 63 network built by TTTMB ligand with Zn(Ⅱ) centers; (c) ABAB packing mode of 2D layers along a axis in complex 2
图2
(a) View of ORTEP of complex 2 with 30% ellipsoid; (b) Two dimensional honeycomb 63 network built by TTTMB ligand with Zn(Ⅱ) centers; (c) ABAB packing mode of 2D layers along a axis in complex 2
Figure2.
(a) View of ORTEP of complex 2 with 30% ellipsoid; (b) Two dimensional honeycomb 63 network built by TTTMB ligand with Zn(Ⅱ) centers; (c) ABAB packing mode of 2D layers along a axis in complex 2
A typical 63 2D topological network is found in complex 2, where each TTTMB ligand coordinates to three Zn(Ⅱ) centers and is considered as a 3-connected nodes. The 2D networks stack in an ABAB packing sequence and further extend to a 3D structure by hydrogen bonds between the methanoic acid and methanol molecules (Table 3).
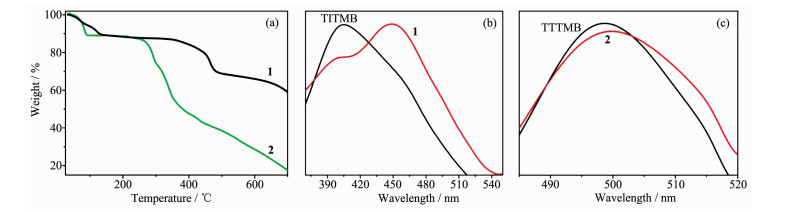 图3
TGA curves of complexes 1 and 2 (a), solid state photoluminescence emission spectra of ligand TITMB and complex 1 with 330 nm excitation (b), ligand TTTMB and complexes 2 with 280 nm excitation (c), respectively
Figure3.
TGA curves of complexes 1 and 2 (a), solid state photoluminescence emission spectra of ligand TITMB and complex 1 with 330 nm excitation (b), ligand TTTMB and complexes 2 with 280 nm excitation (c), respectively
图3
TGA curves of complexes 1 and 2 (a), solid state photoluminescence emission spectra of ligand TITMB and complex 1 with 330 nm excitation (b), ligand TTTMB and complexes 2 with 280 nm excitation (c), respectively
Figure3.
TGA curves of complexes 1 and 2 (a), solid state photoluminescence emission spectra of ligand TITMB and complex 1 with 330 nm excitation (b), ligand TTTMB and complexes 2 with 280 nm excitation (c), respectively
2.3 Thermal stability analyses
Thermogravimetric analyses were performed on grounded samples of 1 and 2 to evaluate their thermal robustness and decomposition behavior and the results are shown in Fig. 3a. Up to 350 ℃, complex 1 undergoes continuous weight loss of 13.1% due to the release of two free methanol and ten coordinated water molecules (Calcd. 13.6%). Obvious rapid weight loss is observed after heating over 350 ℃, which suggests the structure decomposition of complex 1. For complex 2, the first weight loss(11.1%) under 90 ℃ may be assigned to the loss of methanol molecule (5.8%) and partially release of methanoic acid, and the structure could maintained up to 250 ℃.
2.4 Photoluminescence property
The photoluminescence of ligands TITMB, TTTMB, complexes 1 and 2 was studied in the solid state at room temperature. The excitation wavelength for TITMB and 1, TTTMB and 2 is 330 and 280 nm, respectively. The results were depicted in Fig. 3b and 3c.
Complex 1 shows a weak emission band at 400 nm and a strong peak at 450 nm upon the excitation (Fig. 3b). Since free ligand TITMB displays an emission band at 402 nm upon the excitation, the emission band of complex 1 at 400 nm should be due to the ligand-to-ligand (π-π*) charge transfer (LLCT)[22-24]. The 450 nm band should be ascribed to the ligand-to-metal charge transfer (LMCT)[25-27]. Complex 2 exhibits very similar emission band at 498 nm comparing to the free TTTMB ligand (Fig. 3c). Thus, the emission of complex 2 is assigned to be ligand-based (LLCT)[28-29].
3 Conclusions
Two new 2D coordination polymers based on flexible tripodal ligands of TITMB and TTTMB have been synthesized and characterized. In both complexes, the TITMB and TTTMB adopt cis, cis, trans-configur-ation. The counter anions play an important role in the construction of coordination polymer in consid-eration of their different coordination ability, size and so on. In this work, sulfate anions participate in the coordination of the resulted structure of complex 1 as the bridging ligand, while in complex 2, act as terminal coordination ligand only. In addition, complexes 1 and 2 display blue fluorescent emissions in the solid state at room temperature.
-
-
[1]
Feng D W, Jiang H L, Chen Y P, et al. Inorg. Chem., 2013, 52:12661-12667 doi: 10.1021/ic4018536
-
[2]
Zhou H C, Long J R, Yaghi O M. Chem. Rev., 2012, 112:673-674 doi: 10.1021/cr300014x
-
[3]
Liu J, Thallapally P K, McGrail B P, et al. Chem. Soc. Rev., 2012, 41:2308-2322 doi: 10.1039/C1CS15221A
-
[4]
Liu H K, Tan H Y, Liao S, et al. Chem. Commun., 2001:1008-1009
-
[5]
Feng D W, Gu Z Y, Li J R, et al. Angew. Chem. Int. Ed., 2012, 51:10307-10310 doi: 10.1002/anie.201204475
-
[6]
Jiang H L, Feng D W, Wang K C, et al. J. Am. Chem. Soc., 2013, 135:13934-13938 doi: 10.1021/ja406844r
-
[7]
Chen Y, Lykourinou V, Carissa V, et al. J. Am. Chem. Soc., 2012, 134:13188-13191 doi: 10.1021/ja305144x
-
[8]
Wiester M J, Ulmann P A, Mirkin C A. Angew. Chem. Int. Ed., 2011, 50:114-137 doi: 10.1002/anie.v50.1
-
[9]
Chen Y, Lykourinou V, Hoang T, et al. Inorg. Chem., 2012, 51:9156-9158 doi: 10.1021/ic301280n
-
[10]
Liu H K, Sun W Y, Zhu H L, et al. Inorg. Chim. Acta, 1999, 295:129-135 doi: 10.1016/S0020-1693(99)00332-1
-
[11]
徐周庆, 何亚玲, 李慧军, 等.无机化学学报, 2017, 33(5):897-904 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?file_no=20170523
-
[12]
Hiroyasu F, Felipe G, Zhang Y B, et al. J. Am. Chem. Soc., 2014, 136:4369-4381 doi: 10.1021/ja500330a
-
[13]
Reinsch H, Marszalek B, Wack J, et al. Chem. Commun., 2012, 48:9486-9488 doi: 10.1039/c2cc34909d
-
[14]
Yin X J, Zhou X H, Gu Z G, et al. Inorg. Chem. Commun., 2009, 12:548-551 doi: 10.1016/j.inoche.2009.04.012
-
[15]
Henninger S K, Jeremias F, Kummer H, et al. Eur. J. Inorg. Chem., 2012, 16:2625-2634 doi: 10.1002/ejic.201101056
-
[16]
Fan J, Zhu H F, Okamura T, et al. Chem. Eur. J., 2003, 9:4724-4731 doi: 10.1002/chem.v9:19
-
[17]
Fan J, Zhu H F, Okamura T, et al. Inorg. Chem., 2003, 42:158-162 doi: 10.1021/ic020461g
-
[18]
Yan W, Han L J, Jia H L, et al. Inorg. Chem., 2016, 55:8816-8821 doi: 10.1021/acs.inorgchem.6b01328
-
[19]
Nguyen J G, Cohen S M. J. Am. Chem. Soc., 2010, 132:4560-4561 doi: 10.1021/ja100900c
-
[20]
Sheldrick G M. SHELXS-97, Program for X-ray Crystal Structure Solution and Refinement, University of Göttingen, Germany, 1997.
-
[21]
赵仑, 陈瑞战, 王子忱.无机化学学报, 2016, 32(7):1190-1198 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?file_no=20160710
-
[22]
赵越, 翟玲玲, 孙为银.无机化学学报, 2014, 30(1):18-22 http://www.wjhxxb.cn/wjhxxbcn/ch/reader/view_abstract.aspx?file_no=20140110
-
[23]
Hu T P, Xue Z J, Zheng B H, et al. CrystEngComm, 2016, 18:5386-5392 doi: 10.1039/C6CE00674D
-
[24]
Khatua S, Kang J, Huh J O, et al. Cryst. Growth Des., 2010, 10:327-334 doi: 10.1021/cg900917v
-
[25]
Bach S, Panthöfer M, Bienert R, et al. Cryst. Growth Des., 2016, 16:4232-4239 doi: 10.1021/acs.cgd.6b00208
-
[26]
Dong W W, Xia L, Peng Z, et al. J. Solid State Chem., 2016, 238:170-174 doi: 10.1016/j.jssc.2016.03.027
-
[27]
Wang J, Zhang X, Wei Q L, et al. Nano Energy, 2016, 19:222-233 doi: 10.1016/j.nanoen.2015.10.036
-
[28]
Sun D R, Sun F X, Deng X Y, et al. Inorg. Chem., 2015, 54:8639-8643 doi: 10.1021/acs.inorgchem.5b01278
-
[29]
Sun D R, Ye L, Sun F X, et al. Inorg. Chem., 2017, 56:5203-5209 doi: 10.1021/acs.inorgchem.7b00333
-
[1]
-
Figure 1 (a) View of ORTEP drawing of complex 1 with 30% ellipsoid; (b) One dimensional inorganic chain formed by bridging sulfate anion and Cd(Ⅱ); (c) Two dimensional network constructed by TITMB ligands with 1D inorganic chain; (d) AAA stacking modes in complex 1; (e) Cd(H2O)6 units located between the 2D networks further extending the 2D network to 3D framework through hydrogen bonds
Methanol molecule and the hydrogen atoms are omitted for clarity; Symmetry codes: A: x, -1+y, 1+z; B: x, -1+y, z
Table 1. Crystal data and structural refinement of complexes 1 and 2
Complex 1 2 Empirical formula C44H76Cd4N12O28S4 C21H27N9O5Zn Formula weight 1 799.01 550.89 Crystal system Trielinic Triclinic Space group P1 P1 a / nm 1.027 61(11) 0.914 93(9) b / nm 1.233 42(15) 1.259 85(13) c / nm 1.469 65(16) 1.291 34(15) α/(°) 68.646(1) 61.093(1) β/(°) 84.642(2) 84.690(2) γ/(°) 69.495(1) 68.873(1) V / nm3 1.623 7(3) 1.209 4(2) Z 1 2 Dc / (g·cm-3) 1.846 1.513 μ/mm-1 1.51 1.07 F(000) 904 572 GOF 1.028 1.047 Collected reflection 8 251 6 403 Unique reflection 5 587 4 186 R1 0.060 0.093 wR2 0.184 0.235 Table 2. Selected bond lengths (nm) and angles (°) for complexes 1 and 2
Complex 1 Cd1-N9 0.221 4(7) Cd2-N6 0.225 1(7) Cell-01 0.257(2) Cd1-N3 0.224 0(6) Cd2-05 0.234 3(6) Cel 1-09 0.262 0(10) Cd1-O2C 0.225 0(12) Cd2-O10 0.239 1(8) Cd3-012 0.230 8(5) Cd1-O6B 0.239 7(18) Cd3-O13 0.223 4(8) Cd3-O11 0.232 2(6) Cd1-O2 0.262 8(12) N9-Cd1-N3 173.7(3) O6B-Cd1-O2 162.1(5) N3-Cd1-O6B 80.9(5) N9-Cd1-O2C 94.4(4) O6B-Cd1-O1 142.7(7) N3-Cd1-O2C 91.9(4) N9-Cd1-O2 91.6(3) O9-Cd1-O2 119.7(4) N3-Cd1-O9 84.2(3) N9-Cd1-O6B 97.3(5) O9-Cd1-O1 67.0(7) N3-Cd1-O1 90.2(7) N9-Cd1-O9 89.5(3) O9-Cd1-O6B 76.1(5) N3-Cd1-O2 91.8(3) N9-Cd1-O1 87.7(7) O1-Cd1-O2 52.8(7) O2C-Cd1-O6B 104.3(5) O2C-Cd1-O9 176.0(4) O2C-Cd1-O1 112.2(7) O2C-Cd1-O2 59.4(5) N6-Cd2-O5 89.0(2) O13-Cd3-O12 91.9(3) N6-Cd2-O10 84.6(3) O5D-Cd2-O5 180 O13-Cd3-O12A 88.1(3) N6D-Cd2-N6 180 N6-Cd2-O10D 95.4(3) O12-Cd3-O12A 180 N6-Cd2-O5D 91.0(2) O5-Cd2-O10D 90.2(2) O12-Cd3-O11A 92.9(2) O5-Cd2-O10 89.8(2) O12-Cd3-O11 87.1(2) 013A-Cd3-O11 89.2(3) O10D-Cd2-O10 180 O13A-Cd3-O13 180 O13-Cd3-O11 90.8(3) O11A-Cd3-O11 180 Complex 2 Zn1-O3 0.207 2(6) Zn1-N6 0.207 5(6) Zn1-02 0.217 2(8) Zn1-N3 0.209 3(6) Zn1-N9 0.226 1(6) Zn1-O1 0.238 9(8) O3-Zn1-N6 91.1(2) O3-Zn1-N3 91.9(2) O3-Zn1-O1 92.9(3) N6-Zn1-N3 108.0(2) O3-Zn1-O2 91.6(3) N3-Zn1-O1 99.1(3) N6-Zn1-O2 94.8(3) N3-Zn1-O2 156.9(3) N9-Zn1-O1 83.4(2) O3-Zn1-N9 176.2(2) N6-Zn1-N9 91.9(2) N6-Zn1-O1 152.4(3) N3-Zn1-N9 89.5(2) O2-Zn1-N9 85.8(2) O2-Zn1-O1 57.8(3) Symmetry codes: A: 2-x, -y, -z; B: x, 1+y, z; C: 1-x, 2-y, -z; D: -x+2, -y+2, -z+1 for 1. Table 3. Hydrogen bond lengths (nm) and angles (°) for complexes 1~2 Symmetry codes: A: x, -1+y, z; B: 1-x, 1-y, -z for 1.
D-H‧‧‧A d(D-H) / nm d(H‧‧‧A) /nm d(D‧‧‧A) / nm ∠D-H‧‧‧A /(°) 1 O9-H9A‧‧‧O5A 0.085 0 0.195 7 0.278 7(14) 165 O9-H9B‧‧‧O14 0.095 0 0.229 1 0.300(2) 141 O10-H10A‧‧‧O8 0.084 9 0.257 6 0.270(2) 123 O11-H11A‧‧‧O7A 0.085 0 0.214 9 0.299 6(13) 174 O11-H11B‧‧‧O4B 0.085 0 0.204 7 0.277 5(19) 143 O12-H12A‧‧‧O3B 0.085 0 0.191 2 0.274 6(12) 167 O12-H12B‧‧‧O6A 0.085 1 0.212 3 0.296(2) 167 O13-H13C‧‧‧O8A 0.085 0 0.207 9 0.271(2) 130 O14-H14‧‧‧O1 0.082 0 0.223 0 0.284(3) 131 2 O5-H5‧‧‧O1 0.081 6 0.196 2 0.276(2) 166 C6-H6‧‧‧O1 0.093 0 0.253 5 0.308 4(12) 118 C2-H2‧‧‧O4 0.093 0 0.217 3 0.309 5(11) 171 Hydrogen atoms and methanol molecules are omitted for clarity; Symmetry codes: A: 2-x, -y, -z; B: x, 1+y, z; C: 1-x, 2-y, -z; D: 2-x, 2-y, 1-z -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 6
- 文章访问数: 1622
- HTML全文浏览量: 193


 下载:
下载:



 下载:
下载:

