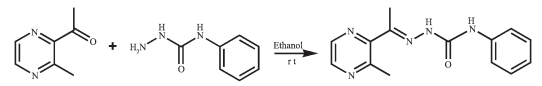
 Figure Scheme 1.
Synthesis route of HL
Figure Scheme 1.
Synthesis route of HL

含吡嗪的缩氨基脲配体Ni (Ⅱ)/Cd (Ⅱ) 配合物的晶体结构及与DNA的相互作用
English
Crystal Structures and DNA Interaction Properties of Ni (Ⅱ) and Cd (Ⅱ) Complexes with a Semicarbazone Ligand Bearing Pyrazine Unit
-
Key words:
- semicarbazone
- / Ni (Ⅱ) complex
- / Cd (Ⅱ) complex
- / pyrazine
- / crystal structure
- / DNA interaction
-
As an important class of ligands, Schiff bases play a crucial role in coordination chemistry and have been widely applied in different fields due to their broad spectrum of biological applications[1-13]. In recent years, the transition metal complexes of Schiff base derivatives bearing a heterocyclic ring (such as pyrrole[2], pyridine[3-6], pyrazine[7-10] and quinoline[11-13]), involving acylhydrazones[3-6], thiosemicarbazones[9-13] and semicarbazones[7-8, 11] have been proved to possess high biological and pharmaceutical activities. Our previous work also shows that two binuclear complexes [Cu2(L)2Cl2] and [Cu2(L)2(OAc)2] with 1-(3-methylpyrazin-2-yl) ethylidene-4-phenylsemicarbazide (HL, Scheme 1) can bind to DNA and have potential pharmaceutical activity[7].
On the other hand, the biological activities of the complexes depend mainly on the metal centers[9-10]. It should be noted that metal-ligand synergism could occur in most cases. In addition, several Ni (Ⅱ) and Cd (Ⅱ) complexes should be investigated because both Ni (Ⅱ) and Cd (Ⅱ) ions are closely related to biochemistry, clinical diagnostics, as well as environmental pollution [8]. Therefore, as the continuation of our work, Ni (Ⅱ) and Cd (Ⅱ) complexes with HL have been synthesized and structurally determined by single-crystal X-ray diffraction. In addition, the interactions between the compounds and ct-DNA have been studied by ethidium bromide (EB) fluorescence probe.
1 Experimental
1.1 Materials and measurements
Solvents and starting materials for synthesis were purchased commercially and used as received. Elemental analysis was carried out on an Elemental Vario EL analyzer. PXRD data were recorded on a Philips X'Pert-MPD instrument with Cu Kα radiation (λ=0.154 056 nm) at 293 K. The IR spectra (ν=4 000 ~400 cm-1) were determined by the KBr pressed disc method on a Bruker V70 FTIR spectrophotometer. The UV spectra were recorded on a Purkinje General TU-1800 spectrophotometer. TG analysis was measured on a Mettler-Toledo TGA/SDTA851e instrument. The interactions between the complexes and ct-DNA are measured using literature method[13] via emission spectra on a Varian CARY Eclipse spectrophotometer.
1.2 Preparations of complexes 1 and 2
The ligand HL was produced by condension of 1-(3-methylpyrazin-2-yl) ethanone and 4-phenylsemicarb-azide following the reported procedure[8]. The complexes 1 and 2 were generated by reaction of the ligand HL (5 mmol) with equimolar of Ni (NO3)2 and CdCl2 in methanol solution (10 mL), respectively. Crystals suitable for X-ray diffraction analysis were obtained by evaporating the corresponding reaction solutions at room temperature.
1: brown blocks. Anal. Calcd. for C29H33N10O3.5Ni (%): C: 54.74; H: 5.23; N: 22.01. Found (%): C: 54.98; H: 5.32; N: 21.86. FTIR (cm-1): ν(N=C-O) 1 615, ν(C=N-N) 1 594, ν(C=N)pyrazine 1 548.
2: colorless needles. Anal. Calcd. for C14H15CdCl2N5O (%): C: 37.15; H: 3.34; N: 15.47. Found (%): C: 37.22; H: 3.16; N: 15.27. FTIR (cm-1): ν(O=C) 1 642, ν(C=N-N) 1 589, ν(C=N)pyrazine 1 544.
1.3 X-ray crystallography
The X-ray diffraction measurement for complexes 1 and 2 were performed on a Bruker SMART APEX Ⅱ CCD diffractometer equipped with a graphite monochromatized Mo Kα radiation (λ=0.071 073 nm) by using φ-ω scan mode. Semi-empirical absorption correction was applied to the intensity data using the SADABS program[14]. The structures were solved by direct methods and refined by full matrix least-square on F2 using the SHELXTL-97 program[15]. All non-hydrogen atoms were refined anisotropically. The occupancy value of O4 atom in complex 1 is 0.5, and thus the H atoms of which are not added. All the other H atoms were positioned geometrically and refined using a riding model. Details of the crystal parameters, data collection and refinements for complexes 1 and 2 are summarized in Table 1.
1 2 Empirical formula CMH33N10O3, Ni C14Hl5CdCl2N50 Formula weight 636.36 452.61 T/K 293(2) 296(2) Crystal system Triclinic Monoclinic Space group P1 P21/c a / nm 1.195 3(19) 0.763 2(3) b / nm 1.199 5(18) 2.043 9(7) c / nm 1.355(2) 1.101 0(4) 109.85(2) 90 α/(°) 100.01(3) 97.741(5) β/(°) 106.07(2) 90 V / nm3 1.677(4) 1.701 8(10) Z 2 4 Dt. / (g·cm-3) 1.260 1.767 Absorption coefficient / mm-1 0.624 1.607 F(000) 666 896 Reflections collected, Unique (Rint) 8 476, 5 852(0.022 3) 8 413, 2 980(0.022 6) Goodness-of-fit (GOF) on F2 1.047 1.043 Final R indices [I > 2σ(I)] R1=0.062 7, wR2=0.194 8 R1=0.034 0, wR2=0.091 7 R indices (all data) R1=0.081 8, wR2=0.211 3 R1=0.039 3, wR2=0.095 5 Table 1. Crystal data and structure refinement for complexes 1 and 2CCDC: 1479242: 1; 1479243: 2.
2 Results and discussion
2.1 Crystal structures description
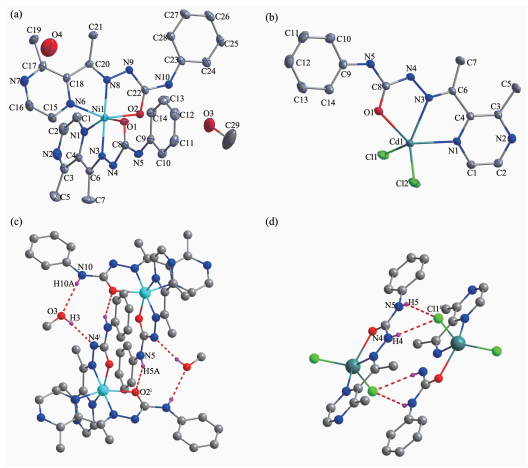
Selected bond distances and angles, hydrogen bonds information for both complexes are listed in Table 2 and 3, respectively. As shown in Fig 1a, the central Ni (Ⅱ) ion in complex 1 is surrounded by two independent anionic ligands with N2O donor set, thus possesses a distorted octahedral coordination geometry: in fact, the N8-Ni1-N3 angle (178.42(13)°) is very close to the theoretical 180°, but the other two, N1-Ni1-O1 and O2-Ni1-N6 are 154.94(14)° and 154.48(13)°, respectively, due to the chelation rings strain. The enolization of C=O bond of the ligand can be confirmed by the bond lengths of C-O being 0.126 6(2) and 0.128 8(1) nm, which are in excellent agreement with previously known semicarbazone complexes[7-8]. The distances of Ni-N/O bonds were in the range of 0.200 9(4)~0.213 3(4) nm, comparable with those in some reported complexes with similar donor set[8]. In the solid state, two complex molecules link with each other into a centrosymmetric moleculardimer via N-H…O hydrogen bonds. In addition, intermolecular N-H…O and N-H…O hydrogen bonds between the complex and crystal methanol molecules are also present (Fig. 1c).
1 Nil-N8 0.200 9(4) Nil-Nl 0.211 4(4) Nil-02 0.213 1(4) Nil-N3 0.201 7(4) Nil-01 0.212 9(4) Nil-N6 0.213 3(4) N8-Nil-N3 178.42(13) Nl-Nil-01 154.94(14) N8-Nil-N6 77.38(17) N8-Nil-Nl 103.30(15) N8-Nil-02 77.10(14) N3-Nil-N6 101.51(17) N3-Nil-Nl 77.82(15) N3-Nil-02 104.00(14) Nl-Nil-N6 92.43(16) N8-Nil-01 101.76(13) Nl-Nil-02 93.62(17) 01-Nil-N6 93.23(15) N3-Nil-01 77.13(12) 01-Nil-02 91.70(16) 02-Nil-N6 154.48(13) 2 Cd1-N3 0.232 7(3) Cd1-Nl 0.237 8(3) Cd1-Cll 0.243 0(13) Cell-01 0.237 6(3) Cd1-C12 0.240 0(13) N3-Cd1-Cll 115.30(9) N3-Crll-01 68.63(10) N3-Cd1-C12 130.98(9) Ol-Cd1-Cll 102.15(9) N3-Cd1-Nl 67.08(10) O1-Cd1-C12 104.82(9) Nl-Cd1-Cll 101.75(9) O1-Crll-Nl 135.28(10) Nl-Cd1-C12 99.37(9) C12-Cd1_Cll 113.54(5) Table 2. Selected bond lengths (nm) and angles (°) in complexes 1 and 2D-H …A d(D-H) / nm d(H …A) / nm d(D …A) / nm ∠D-H…A/(°) 1 O3-H3 …N4ⅰ 0.082 0.200 0.281 5(6) 170.7 N5-H5A…02ⅰ 0.086 0.217 0.297 8(6) 156.0 N10-H10A …O3 0.086 0.230 0.303 1(7) 143.5 2 N5-H5 …Cl1ⅱ 0.086 0.258 0.333 3(4) 146.4 N4-H4 …Cl1Ⅱ 0.086 0.261 0.337 3(4) 148.9 Symmetry codes: ⅰ-x+2, -y, -z; ⅱ1-x, 1-y, 1-z Table 3. Hydrogen bonds information for complexes 1 and 2By contrast, the ratio of the ligand HL and metal is 1: 1, and the semicarbazone acts as a neutral tride-ntate ligand in complex 1 (Fig. 1b), which is confirmed by the double bond nature of C8-O1 (bond length being 0.123 6(4) nm)[7-8]. The Cd (Ⅱ) ion is also coor-dinated with two chloride ions, thus giving a distorted square pyramidal geometry with τ=0.071 7[7]. In the crystal, similar centrosymmetric dimer is formed due to the intermolecular N-H…Cl hydrogen bonds (Fig. 1d).
2.2 PXRD and IR spectra
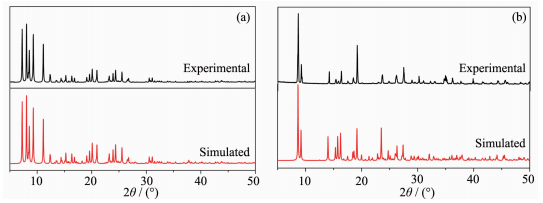
The powder X-ray diffraction (PXRD) for complexes 1 and 2 are presented in Fig. 2, and the experimental pattern is almost same as that of the simulated pattern, except for a little difference about reflection intensities, which manifests that two complexes are the purity phase.
The IR spectral regions for both complexes are more or less similar due to the similarity in coordination modes of the ligand with the metal center. The ν(C=O) of the free ligand is 1 702 cm-1, while it is disappeared in complex 1, meanwhile, new (N=C-O) stretching vibration absorption is observed at 1 615 cm-1, revealing that the C=O in O=C-N moiety has enolizated and the oxygen atom coordinates to the metal ion[7]. By contrast, ν(C=O) of complex 2 shifts to 1 642 cm-1, indicating the coordination of carbonyl group of the neutral ligand HL[8]. The ν(C=N-N) bands of the imine group and pyrazine ring in the ligand HL shift to lower frequency values in the complexes, showing that the N atoms of both units take part in the coordination[7-8]. It is in accordance with the crystal structure study.
2.3 UV spectra
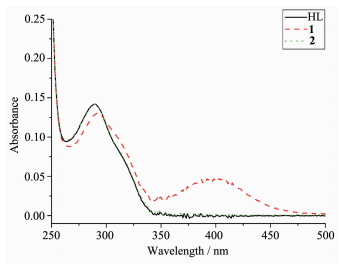
The UV spectra of HL, complexes 1 and 2 in methanol solution (concentration: 1×10-5 mol·L-1) were measured at room temperature (Fig. 3). The spectra of HL features one main band located around 289 nm (ε=14 355 L·mol-1·cm-1), which could be assigned to characteristic π-π* transition of the pyrazine units[8, 11]. The band has almost no change in the spectra of 2 (289 nm, ε=14 312 L·mol-1·cm-1), while slightly red-shifts to 292 nm (ε=13 041 L·mol-1·cm-1) along with hypochromic effect in the spectra of 1. In addition, complex 1 exhibits another band at 400 nm (ε=4 810 L·mol-1·cm-1), corresponding to the ligand-to-metal charge transfer (LMCT)[8]. This indicates that an extended conjugation is formed in anionic ligand after complexation in complex 1.
2.4 Thermal analysis
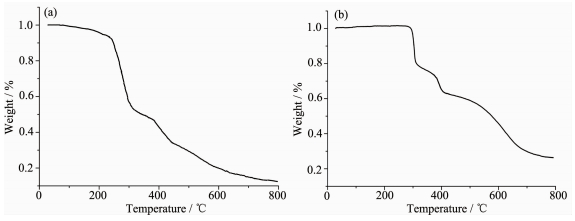
For detecting the thermal stabilities of complexes 1 and 2, thermal gravimetric (TG) analyses were carried out from the room temperature to 800 ℃ with the linear heating rate of 10 ℃·min-1 under argon atmosphere (Fig. 4). The first stage occurs with weight loss of 7.02% below 220 ℃ for complex 1, contributing to the loss of crystal solvent molecules (Calcd. 6.45%). The second and third processes of weight loss appear between 220 to 305 ℃, 305 to 800 ℃, respectively, considered as the decomposition of the two ligands independently. Complex 2 is thermally stable up to about 300 ℃, indicating there exist no solvent molecules in the complex. The weight loss from 300 to 800 ℃ is assigned to the decomposition of the chloride anions and the organic ligand. The remainders of the complexes 1 and 2 might be the metal oxides because the residue weights (12.52% and 26.42%) are agreement with the calculated values of 11.64%, and 28.68%, respectively.
2.4 EB-DNA binding study by fluorescence spectrum
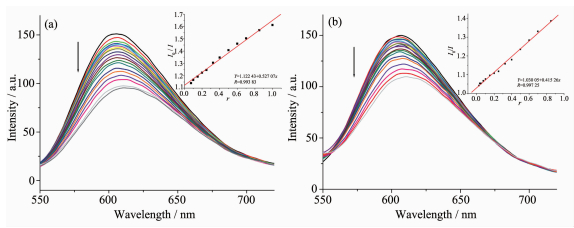
It is well known that EB can intercalate nonspecifically into DNA, which causes it to fluoresce strongly. Competitive binding of other drugs to DNA and EB will result in displacement of bound EB and a decrease in the fluorescence intensity[7, 13]. As shown in Fig. 5, the fluorescence intensities of EB bound to ct-DNA at about 600 nm show remarkable decreasing trends with the increasing concentration of the tested complexes, indicating that some EB molecules are exchanged by the tested complexes. The quenching of EB bound to DNA by the compounds is in agreement with the linear Stern-Volmer equation: I0/I=1+Ksqr[13], where I0 and I represent the fluorescence intensities in the absence and presence of quencher, respectively, Ksq is the linear Stern-Volmer quenching constant, r is the ratio of the concentration of quencher and DNA. In the quenching plots of I0/I versus r, Ksq values are given by the slopes. The Ksq values are 0.527 and 0.415 for complexes 1 and 2, respectively, while that of the ligand HL is tested to be 0.268 in our previous work[7]. The result indicates that interactions of the complexes with DNA are stronger than that of the ligand HL, probably due to the higher rigidity of the complexes, which is in accordance with the literature[7]. In addition, compared with complex 2, the ratio of metal and ligand is 1: 2 and both ligands are deproto-nated and more conjugated in complex 1, thus leading to higher DNA-binding ability. However, the Ksq value of complex 1 is lower than that of the Cu (Ⅱ) complexes with same ligand, namely, [Cu2(L)2Cl2] and [Cu2(L)2(OAc)2] (Ksq values being 0.723 and 0.780, res-pectively)[7]. This is mainly due to the fact that complex 1 has larger sterically hindered effect (octahedron coordination geometry) than the literature complexes (distorted square pyramid coordination geometry)[7], which may prevent the complex 1 molecule from intercalating with DNA.
-
-
[1]
Alagesan L, Bhuvanesh N S P, Dharmaraj N. Dalton Trans., 2013, 42:7210-7223 doi: 10.1039/c3dt50371b
-
[2]
Ye X P, Zhu T F, Wu W N, et al. Inorg. Chem. Commun., 2014, 47:60-62 doi: 10.1016/j.inoche.2014.07.022
-
[3]
陈延民, 解庆范, 刘金花, 等.无机化学学报, 2015, 31(1):74-80 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htmCHEN Yan-Min, XIE Qing-Fan, LIU Jin-Hua, et al. Chinese J. Inorg. Chem., 2015, 31(1):74-80 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htm
-
[4]
Singh P, Singh D P, Singh V P. Polyhedron, 2014, 81:56-65 doi: 10.1016/j.poly.2014.05.066
-
[5]
Recio Despaigne A A, Da Costa F B, Piro O E, et al. Polyhedron, 2012, 38:285-290 doi: 10.1016/j.poly.2012.03.017
-
[6]
Chang H Q, Jia L, Xu Z Q, et al. Inorg. Chem. Commun., 2015, 57:8-10 doi: 10.1016/j.inoche.2015.04.010
-
[7]
林龙, 李先宏, 张波, 等.无机化学学报, 2017, 33(1):143-148 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htmLIN Long, LI Xian-Hong, ZHANG Bo, et al. Chinese J. Inorg. Chem., 2017, 33(1):143-148 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htm
-
[8]
毛盼东, 韩学锋, 吴伟娜, 等.无机化学学报, 2016, 32(1):161-166 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htmMAO Pan-Dong, HAN Xue-Feng, WU Wei-Na, et al. Chinese J. Inorg. Chem., 2016, 32(1):161-166 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htm
-
[9]
Li M X, Zhang L Z, Yang M, et al. Bioorg. Med. Chem. Lett., 2012, 22:2418-2433 doi: 10.1016/j.bmcl.2012.02.024
-
[10]
Li M X, Zhang L Z, Zhang D, et al. Eur. J. Med. Chem., 2011, 46:4383-4390 doi: 10.1016/j.ejmech.2011.07.009
-
[11]
Bourosh P N, Revenko M D, Stratulat E F, et al. Russ. J. Inorg. Chem., 2014, 59:545-557 doi: 10.1134/S0036023614060059
-
[12]
Revenko M D, Bourosh P N, Stratulat E F, et al. Russ. J. Inorg. Chem., 2010, 55:1387-1397 doi: 10.1134/S0036023610090093
-
[13]
毛盼东, 闫玲玲, 王文静, 等.无机化学学报, 2016, 32(3):555-560 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htmMAO Pan-Dong, YAN Ling-Ling, WANG Wen-Jing, et al. Chinese J. Inorg. Chem., 2016, 32(3):555-560 http://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201603027.htm
-
[14]
Sheldrick G M. SADABS, University of Göttingen, Germany, 1996.
-
[15]
Sheldrick G M. SHELX-97, Program for the Solution and the Refinement of Crystal Structures, University of Göttingen, Germany, 1997.
-
[1]
-
Table 1. Crystal data and structure refinement for complexes 1 and 2
1 2 Empirical formula CMH33N10O3, Ni C14Hl5CdCl2N50 Formula weight 636.36 452.61 T/K 293(2) 296(2) Crystal system Triclinic Monoclinic Space group P1 P21/c a / nm 1.195 3(19) 0.763 2(3) b / nm 1.199 5(18) 2.043 9(7) c / nm 1.355(2) 1.101 0(4) 109.85(2) 90 α/(°) 100.01(3) 97.741(5) β/(°) 106.07(2) 90 V / nm3 1.677(4) 1.701 8(10) Z 2 4 Dt. / (g·cm-3) 1.260 1.767 Absorption coefficient / mm-1 0.624 1.607 F(000) 666 896 Reflections collected, Unique (Rint) 8 476, 5 852(0.022 3) 8 413, 2 980(0.022 6) Goodness-of-fit (GOF) on F2 1.047 1.043 Final R indices [I > 2σ(I)] R1=0.062 7, wR2=0.194 8 R1=0.034 0, wR2=0.091 7 R indices (all data) R1=0.081 8, wR2=0.211 3 R1=0.039 3, wR2=0.095 5 Table 2. Selected bond lengths (nm) and angles (°) in complexes 1 and 2
1 Nil-N8 0.200 9(4) Nil-Nl 0.211 4(4) Nil-02 0.213 1(4) Nil-N3 0.201 7(4) Nil-01 0.212 9(4) Nil-N6 0.213 3(4) N8-Nil-N3 178.42(13) Nl-Nil-01 154.94(14) N8-Nil-N6 77.38(17) N8-Nil-Nl 103.30(15) N8-Nil-02 77.10(14) N3-Nil-N6 101.51(17) N3-Nil-Nl 77.82(15) N3-Nil-02 104.00(14) Nl-Nil-N6 92.43(16) N8-Nil-01 101.76(13) Nl-Nil-02 93.62(17) 01-Nil-N6 93.23(15) N3-Nil-01 77.13(12) 01-Nil-02 91.70(16) 02-Nil-N6 154.48(13) 2 Cd1-N3 0.232 7(3) Cd1-Nl 0.237 8(3) Cd1-Cll 0.243 0(13) Cell-01 0.237 6(3) Cd1-C12 0.240 0(13) N3-Cd1-Cll 115.30(9) N3-Crll-01 68.63(10) N3-Cd1-C12 130.98(9) Ol-Cd1-Cll 102.15(9) N3-Cd1-Nl 67.08(10) O1-Cd1-C12 104.82(9) Nl-Cd1-Cll 101.75(9) O1-Crll-Nl 135.28(10) Nl-Cd1-C12 99.37(9) C12-Cd1_Cll 113.54(5) Table 3. Hydrogen bonds information for complexes 1 and 2
D-H …A d(D-H) / nm d(H …A) / nm d(D …A) / nm ∠D-H…A/(°) 1 O3-H3 …N4ⅰ 0.082 0.200 0.281 5(6) 170.7 N5-H5A…02ⅰ 0.086 0.217 0.297 8(6) 156.0 N10-H10A …O3 0.086 0.230 0.303 1(7) 143.5 2 N5-H5 …Cl1ⅱ 0.086 0.258 0.333 3(4) 146.4 N4-H4 …Cl1Ⅱ 0.086 0.261 0.337 3(4) 148.9 Symmetry codes: ⅰ-x+2, -y, -z; ⅱ1-x, 1-y, 1-z -

 扫一扫看文章
扫一扫看文章
计量
- PDF下载量: 4
- 文章访问数: 729
- HTML全文浏览量: 78


 下载:
下载:





 下载:
下载:

